Consequences of genetic selection for environmental impact traits on economically important traits in dairy cows
Purna Kandel A , Sylvie Vanderick A , Marie-Laure Vanrobays A , Hélène Soyeurt A and Nicolas Gengler A BA University of Liege, Gembloux Agro-Bio Tech, TERRA Teaching and Research Center, Passage des Déportés, 2, B-5030 Gembloux, Belgium.
B Corresponding author. Email: nicolas.gengler@ulg.ac.be
Animal Production Science 58(10) 1779-1787 https://doi.org/10.1071/AN16592
Submitted: 2 September 2016 Accepted: 28 April 2017 Published: 20 June 2017
Journal compilation © CSIRO 2018 Open Access CC BY-NC-ND
Abstract
Methane (CH4) emission is an important environmental trait in dairy cows. Breeding aiming to mitigate CH4 emissions require the estimation of genetic correlations with other economically important traits and the prediction of their selection response. In this study, test-day CH4 emissions were predicted from milk mid-infrared spectra of Holstein cows. Predicted CH4 emissions (PME) and log-transformed CH4 intensity (LMI) computed as the natural logarithm of PME divided by milk yield (MY). Genetic correlations of PME and LMI with traits used currently were approximated from correlations between estimated breeding values of sires. Values were for PME with MY 0.06, fat yield (FY) 0.09, protein yield (PY) 0.13, fertility 0.17; body condition score (BCS) –0.02; udder health (UDH) 0.22; and longevity 0.22. As expected by its definition, values were negative for LMI with production traits (MY –0.61; FY –0.15 and PY –0.40) and positive with fertility (0.36); BCS (0.20); UDH (0.08) and longevity (0.06). The genetic correlations of 33 type traits with PME ranged from –0.12 to 0.25 and for LMI ranged from –0.22 to 0.18. Without selecting PME and LMI (status quo) the relative genetic change through correlated responses of other traits were in PME by 2% and in LMI by –15%, but only due to the correlated response to MY. Results showed for PME that direct selection of this environmental trait would reduce milk carbon foot print but would also affect negatively fertility. Therefore, more profound changes in current indexes will be required than simply adding environmental traits as these traits also affect the expected progress of other traits.
Additional keywords: dairy cows, genetic correlation, methane intensity, predicted methane emissions, selection response.
Introduction
The breeding goal in dairy cattle should support the profitability of milk production. Genetic correlations between milk yield (MY) and reproduction, health and fitness traits are negative, and a decline in many functional traits was reported by many studies (Egger-Danner et al. 2015). Accordingly reproduction, health and fitness traits have been included in breeding goal and also selection indices over the past decade. This has resulted in improvement in these traits (Egger-Danner et al. 2015). However, a novel class of traits will need to be considered in the future, those linked to environment concerns. There are at least two major reasons why they are not yet addressed. First direct accurate measurements of these traits on a large scale are difficult to impossible, making their use as selection index traits difficult. Second introduction of environment concerns into breeding goal is also very difficult due to the knowledge gap on how to improve them most efficiently without putting profitability into jeopardy. A major source of the environmental footprint from dairy system is methane (CH4) emissions, which is responsible for 4% of the anthropogenic CH4 emission (FAO 2010). The enteric fermentation in the rumen accounts for a major part of total CH4 emitted from dairy cows. In addition to the environmental impact, CH4 is associated in the literature to a loss of 2–12% of gross energy intake (Johnson and Johnson 1995). Therefore, reducing the CH4 emitted by dairy cows is of both, economic and environmental, interests. Genetic gains are cumulative and small improvements per generation can build over time. To select any new trait, it must have genetic variation and show heritability. Even with currently only limited research available, CH4 traits predicted from milk fatty acids (Kandel et al. 2015) and measured through non-invasive method (Lassen and Løvendahl 2016) have shown sufficient heritability. Previous studies have shown that mid-infrared (MIR) spectroscopy can be used to predict milk fatty acids (Soyeurt et al. 2011) and that milk fatty acids are indirectly related to CH4 emission (Chilliard et al. 2009; Dijkstra et al. 2011). Also, the heritability of MIR milk fatty acids predicted CH4 emission was estimated between 0.21 and 0.40 (Kandel et al. 2015). Moreover, direct prediction of CH4 from MIR spectra without the use of milk fatty acids would be a step forward because by avoiding intermediate steps, prediction errors could be minimised (Gengler et al. 2016). Dehareng et al. (2012) and Vanlierde et al. (2015, 2016) demonstrated that quantification of CH4 emission directly by MIR spectroscopy from milk samples was feasible and can be useful to generate a large number of indirect CH4 phenotypes. Vanlierde et al. (2015) supported by results from Vanrobays et al. (2016) showed that links between CH4 and milk composition are lactation stage specific.
Genetic selection of CH4 emission traits predicted from MIR spectra of milk samples can be imagined because recent research demonstrated genetic variance and sufficient heritability (Kandel et al. 2017). However, the addition of environmental impact traits into the selection goal needs the careful consideration of its impact on other traits in this goal. Before adding any novel traits, additional information about genetic correlations with other objective traits that are already in place and their predicted response are needed. Amongst the correlations needed are those with milk production traits, with functional traits like fertility and with health traits. Udder health (UDH) was represented by somatic cell score (SCS) on a reversed scale. Even if they are not in the breeding objective, correlations to type and body condition scores (BCS), will allow assessing the impact on these traits too.
Therefore, the objective of this study was 2-fold, first to estimate the genetic correlations between environmental impact traits and other traits of interest, and second to quantify their predicted selection response in simple scenarios.
Materials and methods
Genetic valuation of environmental impact traits
Currently no routine genetic evaluation exists in the Walloon region of Belgium for environmental impact traits linked to CH4 emissions. However, in order to approximate genetic correlations among traits, preliminary evaluations were necessary.
Milk samples and prediction of environmental traits
Milk samples were collected from Holstein cows in their first three lactations from January 2010 and March 2014 as routine Walloon milk recording. All milk samples were analysed using a Milkoscan FT6000 spectrometer (Foss, Hillerød, Denmark) by the milk laboratory ‘Comité du Lait’ (Battice, Belgium) to quantify the contents of fat and protein and to record the spectral data. Production records ranged between 5 and 365 days in milk (DIM). Official International Committee of Animal Recording (ICAR) norms were applied. Therefore, observations outside of ranges of 3 –99 kg MY, 1–7% protein content and 1.5–9% fat content were not used for the calculations as suggested in these norms (ICAR 2016).
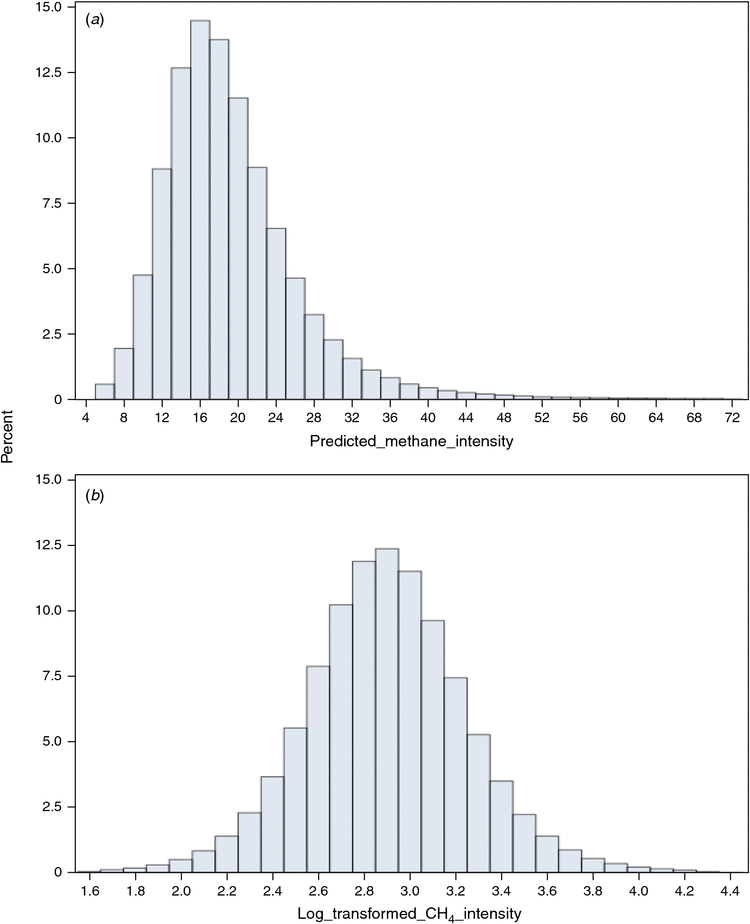
The CH4 emission (PME; g/day) was predicted from the recorded and standardised (Grelet et. al. 2015) milk MIR spectral database of Walloon milk recording using the equation developed by Vanlierde et al. (2015). The predicted CH4 intensity (PMI; g/kg of milk) was defined as the ratio of PME divided by the total milk MY recorded for the considered test-day. The distribution of PMI was non-normal and skewed therefore (Fig. 1) presenting a log-normal aspect. Therefore, PMI was log-transformed and called log-transformed CH4 intensity (LMI) using the natural logarithm. The datasets of predicted environmental traits had 700 505 test-day records from 58 412 first three parity cows sired by 2455 bulls. The heritabilities of PME and LMI were estimated to be 0.25 and 0.18 respectively (Kandel et al. 2017). Within cow, if parity 3 was present, parities 1 and 2 had to be present, and if parity 2 was present, parity 1 had to be present. Animals which had, based on their pedigree, at least 75% of confirm Holstein genetics in their breed composition were kept for this study. Pedigree data were extracted from pedigree used for routine Walloon genetic evaluation and contained 119 068 animals born after 1990, which permitted pedigree up to three generations back.
|
Model
A single trait multiple lactation random regression test-day model was used to estimate the genetic parameters and breeding values of each of PME and LMI. The model can be presented as follows:
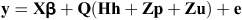
where y was the vector of observations for each trait (PME or LMI), β was the vector of fixed effects (herd × test-day, days in milk (24 classes of 15 days’ interval), and age at calving (9 classes: 21–28 months, 29 to 32 months, and 33 months and more for first lactation; 31–44 months, 44–48 months, and 49 months and more for second lactation and 41–57 months, 57–60 months, and 60 months and more for third lactation), h was the vector of random within-herd lactation curve effects, p was the vector of permanent environmental (PE) random effects, u was the vector of additive genetic effects; Q was the matrix containing the coefficients of 2nd order Legendre polynomial regressors; e was the vector of residuals; X was an incidence matrix assigning observations to levels of fixed effects., H and Z were incidence matrices assigning regressors to random regression coefficients.
Variance components and solutions of mixed model equations
The variance components were estimated by Bayesian method with Gibbs sampling. Priors of variance components were estimated using univariate models using the average information REML. Posterior means of (co)variance components were calculated using 90 000 samples after a burn-in of 10 000 samples. The estimated breeding values (EBV) were calculated using a BLUP approach using obtained variance components.
Economically important traits
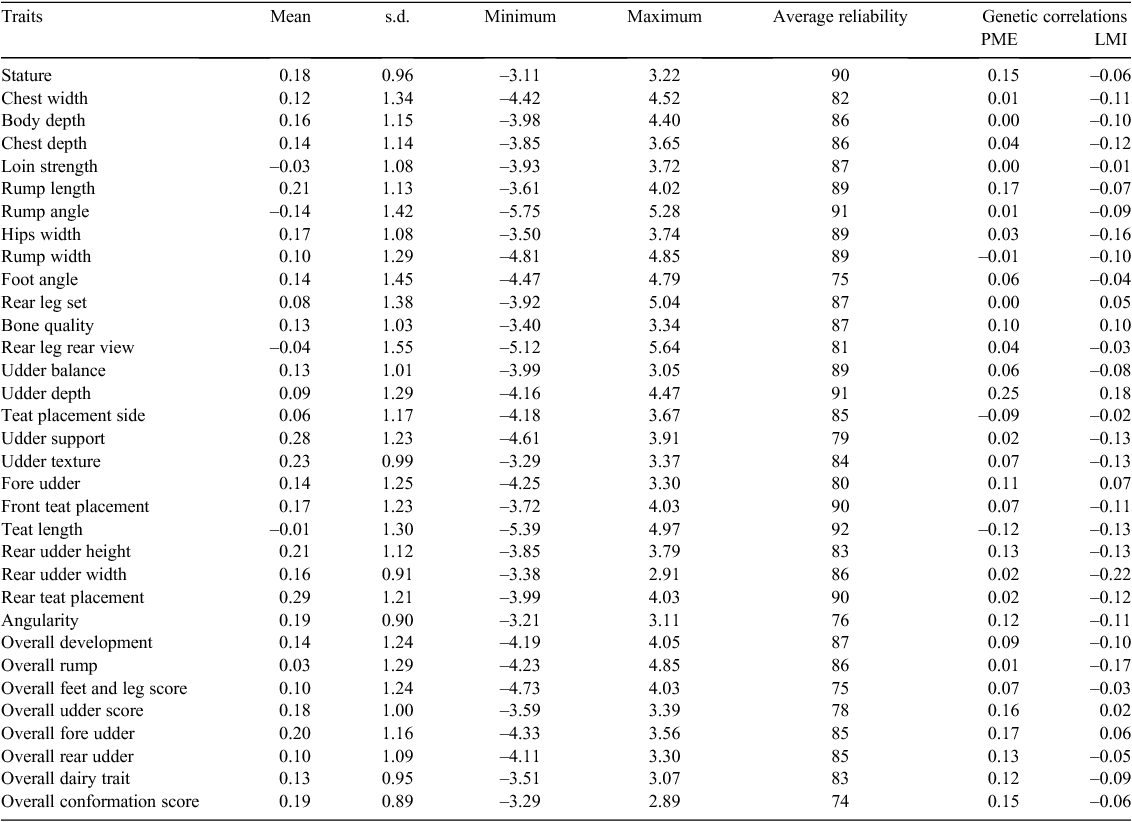
The Walloon Breeding Association (Ciney, Belgium) uses for Holstein dairy cows a selection index called V€G (Vanderick et al. 2015). Table 1 gives the relative importance of the different traits used in the current index. This index was obtained to select for a breeding goal that was derived based on a lifetime economic function including production and functional traits (N. Gengler, pers. comm.). The three categories of traits under routine genetic evaluation in Wallonia and included in the selection index are production, functional traits and type traits. The later were not considered having an economic value on their own, but contributing to the traits in the breeding goal (N. Gengler, pers. comm.). Production traits included MY, fat yield (FY) and protein yield (PY), functional traits were UDH and longevity and more recently fertility and calving traits. These traits were (a) combined female fertility (CFF), (b) direct calving ease (DCE) and (c) maternal calving ease (MCE). The genetic correlations were calculated for all fertility-related traits however response to selection was only calculated for combined female fertility. CFF representing pregnancy rate and higher values are better. Direct calving ease and maternal calving ease were just recently added in selection index; therefore, the responses were not calculated in this study however genetic correlations were calculated. The trait BCS is currently used in computations of EBV for combined fertility and not directly in the index or even breeding goal. However, there are indications (e.g. Vanrobays et al. 2016) that CH4 production through its links to fatty acids and intake interacts with body fat mobilisation. Effects of selection on CH4 were also computed for BCS, an indicator of body fat mobilisation and, indirectly, an important element for a long-term effect on fertility. Longevity was also calculated from direct longevity trait plus genetically correlated type traits. In addition to production and functional traits, a total of 33 type traits (recorded and derived) are also part of Walloon animal genetic evaluation system. Type traits were broadly classified as body capacity, udder and feet and leg traits. Details of all these traits definitions, their genetic model for parameter estimations are described in Vanderick et al. (2015) and Croquet et al. (2006). The EBV of sires for production, functional and type traits were extracted from the database containing the EBV computed for the official Walloon genetic evaluation 2016 March run. These EBV were of domestic, but more often of Multiple Across Country Evaluation (MACE) origin, provided by the INTERBULL Center (Uppsala, Sweden).
Approximated genetic correlations
Very few genetic evaluation systems are completely multivariate across all the index traits. An implication of this is also that genetic correlations amongst these traits are not known. In order to achieve the objective of this study approximated values were needed. Pearson’s correlations among EBV of sires were computed in order to get lower-bound estimates of genetic correlations. The productive life of higher yielding cows goes over 2.5 to 3.2 parities before being culled (e.g. Hare et al. 2006). As life (3 parities) genetic correlations and selection response is easy to understand and interpret, therefore in this study we studied all traits cumulated over life time (at least 3 parities). A total of 2455 bulls had daughters with environmental records and subsequently EBV. These EBV were centred and expressed as average daily values but based on cumulative 305-day emissions over the three lactations (Table 2). For this 2455 sires the corresponding sire EBV for current official genetic evaluations were selected when they showed sufficient reliability (limits depending upon the traits: 50–99% for production traits and 25–99% for functional and type traits). Table 2 gives the figures of selected bulls ranging from 1369 to 1427 for production and functional traits. The equivalent figure was 1422 sires for type traits.
Selection scenarios and predicted responses
Five selection scenarios were proposed to calculate the selection response. Scenario I was the current Walloon selection index V€G (status quo), and from second to fifth selection scenarios were 5%, 12.5% and 25% and 50% addition of CH4 emission traits (PME respectively LMI) and proportional reduction on other traits present in current index (Table 1). The weight of CH4 traits were put negative because we were interested to reduce the CH4 emission from our dairy production. Relative genetic changes for each trait from selection based on these alternative total indexes were estimated as r = bʹG where r = vector of relative genetic gain on all traits; and b = vector of proportional index weights; G = matrix of genetic correlations between index traits and goal traits. As only relative changes were relevant for this study, selection intensity was set to 1 and response was calculated for one generation.
Results
Environmental traits and economic important traits descriptions
The average ± s.d. PME was 443.86 ± 77.04 (g/day) and LMI was 2.87 ± 0.36 for first three lactations. The sire EBV of CH4 emissions traits that had daughters in production were accumulated over three parities, expressed on a daily basis and presented in Table 2. Similarly the corresponding sire EBV obtained from official Walloon genetic evaluation for production (MY, FY and PY) and functional traits (Fertility, BCS, UDH and longevity) were also presented in Table 2. Average reliabilities of selected groups ranged between 61 (for maternal calving ease) and 91 (for UDH). The selected sire EBV for type traits are presented in Table 3 with average reliabilities between 74 and 91.
Genetic correlations between environmental traits and economic important traits
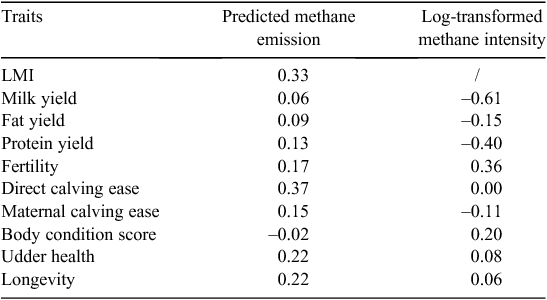
The approximate genetic correlations based on correlation between sire EBV, hereafter called for simplicity genetic correlation, between PME and LMI and production and functional traits are presented in Table 4. The genetic correlation between PME and LMI was estimated 0.33. PME had small positive genetic correlations with milk production traits i.e. 0.06 with MY, 0.09 with FY and 0.13 with PY. However, the genetic correlations between LMI and milk production traits were negative and in case with MY was highly negative (–0.61) and moderate negative with PY (–0.40) and low negative (–0.15) with FY. The genetic correlation of combined female fertility with both CH4 traits was positive but higher in case of LMI (0.36 vs 0.17). Other reproductive traits (DCE and MCE) also had positive genetic correlation with PME however negative correlation were observed between LMI and MCE. The correlation between PME and BCS was very close to zero but 0.20 between LMI and BCS. UDH had positive genetic correlation with both CH4 traits. Finally, longevity had positive genetic correlation with both CH4 traits (Table 4).
|
The genetic correlations between CH4 traits and type traits are reported in Table 3. The genetic correlation between PME and 33 type traits ranged from –0.12 to 0.25 and between LMI and 33 type traits ranged from –0.22 to 0.18. The body capacity traits also had in general positive genetic correlations with PME and negative genetic correlations with LMI. The bodyweight related traits like stature and angularity had positive genetic correlations with PME and negative genetic correlations with LMI. The udder capacity traits also had in general positive genetic correlation with PME and negative genetic correlations with LMI.
Expected genetic changes under selection scenarios
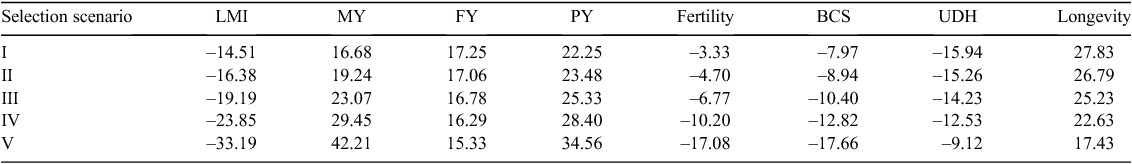
The selection response to each scenario of selecting PME is reported in Table 5. The PME would be increased by 2% without selecting this trait but through correlated responses of other traits. A relative weight of 12.5% on PME (selection scenario III) was necessary to decrease PME. A relative weight of 25% of PME (selection scenario IV) generated a response of PME by –6%, MY by 15%, FY by 6%, PY by 11%, fertility by –4%, BCS by –11%, UDH by –13% and longevity by 22%. In all scenarios MY, FY and PY also increased except with the extreme selection scenario V (50% weight on PME), which decreased FY and PY. In all PME reduction scenarios, fertility, BCS and UDH would decrease. Given that the longevity has currently a very high weight in Walloon index (~21%) longevity has a very positive response in selection scenario I, however the progress would be reduced with each scenario selecting for lower PME.
The favourable genetic gain would be achieved for LMI in all selection scenarios (Table 6). The expected response of LMI would range from –15% to –33% from selection scenario I to selection scenario V. MY, FY and PY would increase in each scenario. For example by the addition of 25% of LMI, the resulting response would be for LMI by –24%, MY by 29%, FY by 16%, PY by 28%, fertility by –10%, BCS by –13%, UDH by –13% and longevity by 23%.
Discussion
The final objective of this study was to assess the response of selection for environmental traits by selecting them directly as well as the correlated responses of other economic important traits. Similarly, the motivation was also to improve the understanding of the genetic influence and their correlations on CH4 emission by dairy cows. Currently, there is no direct economic incentive for a dairy producer to develop a program which reduces CH4 emissions. Given that increasing significance of climate change, in national agendas but also for the dairy industry, environmental traits would need to be included in dairy cattle breeding. Similarly, societal demands are changing from both environmental and economic perspectives and CH4 emission traits could be added in the breeding goals defined for dairy cows in the near future (Hayes et al. 2013).
All of genetic correlations between production, functional and type traits and CH4 emissions traits were revolving around the efficiency and inefficiency of animal from intake, digestion, production, reproduction and survival. Dairy cows seem to partition first energy for production, then for reproduction and finally for survival. More efficient dairy cows will produce more milk relative to the amount of feed ingested and less energy lost as CH4. All production traits had small positive correlations with PME and high negative correlations with LMI, as was expected given its definition. The very negligible positive genetic correlation of PME with production traits suggested that these traits are not able to predict CH4 emissions alone in dairy cows on a genetic level. Similarly, positive genetic correlations observed between PME and fat and protein corrected MY (0.07 ± 0.09) (Lassen and Løvendahl 2016) was similar to this study (0.06 with MY). There is ongoing debate on those figures because they appear low, but one should not forget that even if PME is also driven by intake it is also strongly related to energy lost or energy efficiency, a different mechanism a priori not (strongly) linked to intake. On a phenotypical level our recent research (P. Kandel, unpubl. data) showed that with increasing MY, the correlation with PME is decreasing and eventually inversing as always higher producing animals produce more and more from body reserve mobilisation than intake. In beef cattle, using small preliminary analysis using genomic selection, response to selection of CH4 yield (CH4/kg dry matter intake) was estimated to be reduction by 4% in 10 years (Hayes et al. 2016). In dairy cows, using prediction from feed intake, PME would at least theoretically decrease in the order of 11–26% in 10 years (de Haas et al. 2011). However, those predictions were totally different than prediction used in this study as they assumed that the major driving factor behind PME were only intake driven, therefore, direct comparison was difficult.
The reduction of LMI by 15% through the current Walloon selection index was similar to results obtained by Bell et al. (2011). These authors demonstrated that genetic selection for energy-corrected milk reduced CH4/energy-corrected milk, (which is similar to LMI) by 15% for the first three lactations until mature size and maximum MY are achieved. Moreover, increasing selection pressure for reduced LMI gives a strong positive reaction of MY and associated traits. Therefore, as expected from these results, the functional traits would have negative to strongly negative correlated response. Fertility and BCS would be mostly affected but also longevity.
The genetic correlation between environmental traits and fertility could indicate that more resource inefficient cows show better female fertility and therefore simultaneous selection for both traits might be difficult. However, a breeding strategy emphasising female fertility traits would improve cow fertility and reduce within-herd replacement rates and consequently reduced replacements contribute to decreasing CH4 emissions in herd level (Knapp et al. 2014) but the relationship in individual level is not known yet.
The BCS would decrease in all scenarios of selection on either PME or LMI. The substantial genetic correlation between BCS and LMI reduced BCS and positive genetic correlation between BCS and fertility (Bastin et al. 2012) had led to reduction in both traits. It is also well known that the early lactation period is characterised by body fat mobilisation, negative energy balance (van Knegsel et al. 2007), which is also related to CH4 emissions, so test-day genetic correlations are more important than average of whole lactations.
The fact that longevity had also positive genetic correlations with emissions could indicate that the higher CH4-producing cows might be more efficient in survival. However, like for improved fertility, by promoting longevity emissions from replacement would be diluted. In addition their effect in individual level of emission is unresolved (Grandl et al. 2016).
In sheep, it was demonstrated that smaller body confirmation animals had smaller rumen and shorter duration of ruminal passage, which leads to less CH4 (Goopy et al. 2014). In this study, almost all capacity and body size-related traits like stature, chest width, rump length and angularity had positive genetic correlations with PME, which suggested increased body capacity and bodyweight increased also PME. However, the body capacity type traits had negative genetic correlations with LMI, suggesting that selection for LMI would preserve these traits.
Even without selection on LMI the reduction in CH4 intensity was already substantial due to the negative correlation with production traits. The speed of reduction would be faster if we add this new trait to the selection index, however the decrease in fertility would be substantial unless fertility traits were also added or their weight increased in the selection index.
This study has some limitations. First, the analyses were only based on correlations of sire EBV. A more direct method would be to estimate genetic correlations from the data using appropriate, e.g. bivariate, models. However, such approach would have required variance components estimation for a great number of bivariate models including complicated, e.g. random regression, models. Therefore, for this study, approximations were used and as presented by Calo et al. (1973), correlations between breeding values do not fully reflect the genetic relationships between two traits and they might underestimate them. Second, a better approach to create a selection index would be to put appropriate economic weights to environmental traits instead of adding a linear percentage in selection scenarios. However, even if there is an economic value of CH4 emission in the industrial sector, this is not yet the case in agriculture. An alternative strategy would be to optimise expected gains, developing weights retrospectively. Third, the responses presented in Tables 5 and 6 assume that all breeding values for all traits have equal reliability. That might not be the case at the moment of selection. It is therefore somewhat idealised scenarios but in practice accuracy will differ due to heritability and different recording (e.g. longevity and fertility). However, this study showed practical significance of current selection and its effect on PME and LMI where PME is increasing but CH4 intensity decreasing.
Conclusions
This study presented novel results. First, under the hypothesis to continue using the current Walloon index, without directly selection for environmental traits, PME would be increased but LMI would be decreased through correlated responses to the selection for correlated traits. This is the expected result that gains are currently only achieved per unit produced. Second, by giving direct selection pressure on environmental traits, they would respond to selection, but would also change fundamentally the responses in other traits. These responses were quantified in various scenarios. One of the scenarios – reducing all traits weight by 25% of current index and addition of 25% of PME would reduce gains in FY and PY and almost all functional traits (fertility, BCS, and longevity) would need to be protected. The addition of 25% of LMI would shift the emphasis on production traits, especially MY, and affect even stronger functional traits. In conclusion, direct selection of environmental traits would reduce milk carbon foot print but more profound changes in current indexes will be required than simply adding environmental traits, as adding these traits to the selection index would affect the equilibrium between the other traits.
Acknowledgements
The authors acknowledge the support provided by the European Commission under the Seventh Framework Program for the GreenhouseMilk and GplusE projects, Grant Agreements FP7-PEOPLEITN-2008 238562 and FP7-KBBE-613689, the contribution of the COST Action FA1302 Methagene (http://www.methagene.eu, accessed 12 May 2017) and of the Public Service of the Walloon Region of Belgium (SPW-DGO3) through the Methamilk project. The content of the paper reflects only the view of the authors; the European Community is not liable for any use that may be made of the information contained in this publication. The authors acknowledge the Walloon Breeding Association (Ciney, Belgium) for providing the milk recording and pedigree databases and the Comite du Lait (Battice, Belgium) for the supplying of the MIR spectra. The used MIR prediction equation was developed in a collaborative effort by the Walloon Agricultural Research Center (Gembloux, Belgium). Computational resources have been provided by the Plateau de Calcul et Modélisation informatique (CAMI) of the University of Liège, Gembloux Agro-Bio Tech and the Consortium des Equipements de Calcul Intensif (CECI) of the Federation Wallonia-Brussels (Brussels, Belgium), funded by the National Fund for Scientific Research (Brussels, Belgium) funded under grant 2.5020.11.
References
Bastin C, Berry DP, Soyeurt H, Gengler N (2012) Genetic correlations of days open with production traits and contents in milk of major fatty acids predicted by mid-infrared spectrometry. Journal of Dairy Science 95, 6113–6121.| Genetic correlations of days open with production traits and contents in milk of major fatty acids predicted by mid-infrared spectrometry.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVCnsb7K&md5=1f366ce337e103e8c469deb95d27a70fCAS |
Bell MJ, Wall E, Simm G, Russell G (2011) Effects of genetic line and feeding system on methane emissions from dairy systems. Animal Feed Science and Technology 166–167, 699–707.
| Effects of genetic line and feeding system on methane emissions from dairy systems.Crossref | GoogleScholarGoogle Scholar |
Calo LL, McDowell RE, Van Vleck LD, Miller PD (1973) Genetic aspects of beef production among Holstein–Friesians pedigree selected for milk production. Journal of Animal Science 37, 676–682.
| Genetic aspects of beef production among Holstein–Friesians pedigree selected for milk production.Crossref | GoogleScholarGoogle Scholar |
Chilliard Y, Martin C, Rouel J, Doreau M (2009) Milk fatty acids in dairy cows fed whole crude linseed, extruded linseed, or linseed oil, and their relationship with methane output. Journal of Dairy Science 92, 5199–5211.
| Milk fatty acids in dairy cows fed whole crude linseed, extruded linseed, or linseed oil, and their relationship with methane output.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtF2qtrfE&md5=c383c72f671a923509af27037ee83b37CAS |
Croquet C, Mayeres P, Gillon A, Vanderick S, Gengler N (2006) Inbreeding depression for global and partial economic indexes, production, type, and functional traits. Journal of Dairy Science 89, 2257–2267.
| Inbreeding depression for global and partial economic indexes, production, type, and functional traits.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xlt1eqsb4%3D&md5=375046a0036c9ba895e1045169145ad6CAS |
de Haas Y, Windig YJJ, Calus MPL, Dijkstra J, de Haan M, Bannink A, Veerkamp RF (2011) Genetic parameters for predicted methane production and potential for reducing enteric emissions through genomic selection. Journal of Dairy Science 94, 6122–6134.
| Genetic parameters for predicted methane production and potential for reducing enteric emissions through genomic selection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsFClsbvL&md5=416eec2c2af88d1243ae3daaceb32ca2CAS |
Dehareng F, Delfosse C, Froidmont E, Soyeurt H, Martin C, Gengler N, Vanlierde A, Dardenne P (2012) Potential use of milk mid-infrared spectra to predict individual methane emission of dairy cows. Animal 6, 1694–1701.
| Potential use of milk mid-infrared spectra to predict individual methane emission of dairy cows.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1Gis7zP&md5=ed22c4f580dd9b9fe19071c9ad8b0a9aCAS |
Dijkstra J, van Zijderveld SM, Apajalahti JA, Bannink A, Gerrits WJJ, Newbold JR, Perdok HB, Berends H (2011) Relationships between methane production and milk fatty acid profiles in dairy cattle. Animal Feed Science and Technology 166–167, 590–595.
| Relationships between methane production and milk fatty acid profiles in dairy cattle.Crossref | GoogleScholarGoogle Scholar |
Egger-Danner C, Cole JB, Pryce JE, Gengler N, Heringstad B, Bradley A, Stock KF (2015) Invited review: overview of new traits and phenotyping strategies in dairy cattle with a focus on functional traits. Animal 9, 191–207.
| Invited review: overview of new traits and phenotyping strategies in dairy cattle with a focus on functional traits.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtFyktLs%3D&md5=ba2655cf3c430c7356c124deaf2d20d4CAS |
FAO (2010) ‘Greenhouse gas emissions from the dairy sector: a life cycle assessment.’ Prepared by P Gerber, T Vellinga, C Opio, B Henderson, H Steinfeld. (FAO: Rome, Italy)
Gengler N, Soyeurt H, Dehareng F, Bastin C, Colinet FG, Hammami H, Vanrobays ML, Lainé A, Vanderick S, Grelet C, Vanlierde A, Froidmont E, Dardenne P (2016) Capitalizing on fine milk composition for breeding and management of dairy cows. Journal of Dairy Science 99, 4071–4079.
| Capitalizing on fine milk composition for breeding and management of dairy cows.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XktVSlug%3D%3D&md5=2f93e72ba1804bfe2d36966707497665CAS |
Goopy JP, Donaldson A, Hegarty R, Vercoe PE, Haynes F, Barnett M, Oddy VH (2014) Low-methane yield sheep have smaller rumens and shorter rumen retention time. British Journal of Nutrition 111, 578–585.
| Low-methane yield sheep have smaller rumens and shorter rumen retention time.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXisVCgtL8%3D&md5=1628caf257355975b0c363fd4bd93f7dCAS |
Grandl F, Amelchanka SL, Furger M, Clauss M, Zeitz JO, Kreuzer M, Schwarm A (2016) Biological implications of longevity in dairy cows: 2. Changes in methane emissions and efficiency with age. Journal of Dairy Science 99, 3472–3485.
| Biological implications of longevity in dairy cows: 2. Changes in methane emissions and efficiency with age.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XjsVamsrk%3D&md5=9ae7aba62102fcc77e5a4697361de718CAS |
Grelet C, Fernández Pierna JA, Dardenne P, Baeten V, Dehareng F (2015) Standardization of milk mid-infrared spectra from a European dairy network. Journal of Dairy Science 98, 2150–2160.
| Standardization of milk mid-infrared spectra from a European dairy network.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXis1eitr0%3D&md5=bed5cf000990c702aa4faedfa2d7da97CAS |
Hare E, Norman HD, Wright JR (2006) Survival rates and productive herd life of dairy cattle in the United States. Journal of Dairy Science 89, 3713–3720.
| Survival rates and productive herd life of dairy cattle in the United States.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xos1Kht7k%3D&md5=1959ffe96c09c567e4d62f25356e3c4eCAS |
Hayes BJ, Lewin HA, Goddard ME (2013) The future of livestock breeding: genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends in Genetics 29, 206–214.
| The future of livestock breeding: genomic selection for efficiency, reduced emissions intensity, and adaptation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvVKqurbM&md5=6e692580551c51331286b4cf55f236d3CAS |
Hayes BJ, Donoghue KA, Reich CM, Mason BA, Bird-Gardiner T, Herd RM, Arthur PF (2016) Genomic heritabilities and genomic estimated breeding values for methane traits in Angus cattle. Journal of Animal Science 94, 902–908.
| Genomic heritabilities and genomic estimated breeding values for methane traits in Angus cattle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhsVCks7bN&md5=e47d8131c8def0879c2b705d75a43446CAS |
ICAR (2016) ‘International agreement of recording practices: ICAR recording guidelines.’ (ICAR: Rome) Available at http://www.icar.org/wp-content/uploads/2016/03/Guidelines-Edition-2016.pdf [Verified 12 February 2017]
Johnson KA, Johnson DE (1995) Methane emissions from cattle. Journal of Animal Science 73, 2483–2492.
| Methane emissions from cattle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXnsVCntb8%3D&md5=66b17b1df98cdfa8f6b620f3937fcbffCAS |
Kandel PB, Gengler N, Soyeurt H (2015) Assessing variability of literature based methane indicators traits in a large dairy cow population. Biotechnologie, Agronomie, Société et Environnement 19, 11–19.
Kandel PB, Vanrobays ML, Vanlierde A, Dehareng F, Froidmont E, Gengler N, Soyeurt H (2017) Genetic parameters for predicted methane emission traits and their relationship with milk production traits in Holstein cows. Journal of Dairy Science
| Genetic parameters for predicted methane emission traits and their relationship with milk production traits in Holstein cows.Crossref | GoogleScholarGoogle Scholar |
Knapp JR, Laur GL, Vadas PA, Weiss WP, Tricarico JM (2014) Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. Journal of Dairy Science 97, 3231–3261.
| Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXmtlCnt74%3D&md5=5a9085b73aa64f0be667c79bfe684bd5CAS |
Lassen J, Løvendahl P (2016) Heritability estimates for enteric methane production in dairy cattle using non-invasive methods. Journal of Dairy Science 99, 1959–1967.
| Heritability estimates for enteric methane production in dairy cattle using non-invasive methods.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhsVKhsbo%3D&md5=89517241141358a911f484aac664f756CAS |
Soyeurt H, Dehareng F, Gengler N, McParland S, Wall E, Berry DP, Coffey M, Dardenne P (2011) Mid-infrared prediction of bovine milk fatty acids across multiple breeds, production systems, and countries. Journal of Dairy Science 94, 1657–1667.
| Mid-infrared prediction of bovine milk fatty acids across multiple breeds, production systems, and countries.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXnvFChtro%3D&md5=d51f9d6b983929ba4608d504ba453b18CAS |
van Knegsel ATM, Van denBrand H, Dijkstra J, Van Straalen WM, Heetkamp MJ, Tamminga S, Kemp B (2007) Dietary energy source in dairy cows in early lactation: energy partitioning and milk composition. Journal of Dairy Science 90, 1467–1476.
| Dietary energy source in dairy cows in early lactation: energy partitioning and milk composition.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXitlWnu7g%3D&md5=a89c39c1310d925b7985cfb15429d286CAS |
Vanderick S, Bastin C, Gengler N (2015) Description of the genetic evaluation systems used in the Walloon Region of Belgium. Available at http://www.elinfo.be/docs/GESen1508.pdf [Verified 12 February 2017]
Vanlierde A, Vanrobays M-L, Dehareng F, Froidmont E, Soyeurt H, McParland S, Lewis E, Deighton MH, Grandl F, Kreuzer M, Gredler B, Dardenne P, Gengler N (2015) Hot Topic: Innovative lactation stage dependent prediction of methane emissions from milk mid-infrared spectra. Journal of Dairy Science 98, 5740–5747.
| Hot Topic: Innovative lactation stage dependent prediction of methane emissions from milk mid-infrared spectra.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXpt1Sksrs%3D&md5=3922fe283ad0cfa7f12c262aa543eddeCAS |
Vanlierde A, Vanrobays M-L, Gengler N, Dardenne P, Froidmont E, Soyeurt H, McParland S, Lewis E, Deighton MH, Mathot M, Dehareng F (2016) Milk mid-infrared spectra enable prediction of lactation-stage dependent methane emissions of dairy cattle within routine population-scale milk recording schemes. Animal Production Science 56, 258–264.
| Milk mid-infrared spectra enable prediction of lactation-stage dependent methane emissions of dairy cattle within routine population-scale milk recording schemes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xis1ams7s%3D&md5=8d2e869dff97a2a1eabd00391448d7fbCAS |
Vanrobays M-L, Bastin C, Vandenplas J, Hammami H, Soyeurt H, Vanlierde A, Dehareng F, Froidmont E, Gengler N (2016) Changes throughout lactation in phenotypic and genetic correlations between methane emissions and milk fatty acid contents predicted from milk mid-infrared spectra Journal of Dairy Science 99, 7247–7260.
| Changes throughout lactation in phenotypic and genetic correlations between methane emissions and milk fatty acid contents predicted from milk mid-infrared spectraCrossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhtVymsr3P&md5=da00ad82988284f0e5b8aea67dc16918CAS |