Development of an algorithm to correlate Physi-Trace pig liver data with pork meat data
R. J. Watling A , H. A. Channon B and G. S. H. Lee A CA Food Science Solutions, Gwelup, WA 6018.
B Australian Pork Limited, Barton, ACT 2600.
C Corresponding author. Email: gshlee@bigpond.com
Animal Production Science 57(12) 2490-2490 https://doi.org/10.1071/ANv57n12Ab124
Published: 20 November 2017
Physi-Trace was designed to facilitate the provenance of pork back to a point of origin. Throughout Physi-Trace, it has been shown that it is possible to validate whether a pork product is Australian or not and to establish the origin (farm/tattoo) of Australian pork. This has been possible through continued sampling of known origin pork meat and the subsequent trace elemental analysis and statistical interpretation of the resulting data. A system is now in place that will facilitate the traceability of fresh Australian pork. Associated with the traceability of fresh pork is the traceability of pork offal. Preliminary research by Kreitals (2013) indicated that it was possible to link the elemental profile of offal varieties to the elemental profiles of fresh pig meat using several algorithms thus allowing the origin determination of pig offal to be conducted using the Physi-Trace framework. The algorithms developed by Kreitals (2013) were limited to a single processor and required further research to verify their robustness. The objective of this research was to undertake a detailed study to develop a universal algorithm that could be used to trace analytical data for liver back to their equivalent pork meat data and establish region of origin.
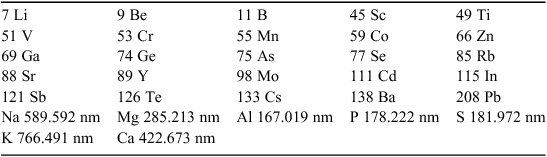
The experiment required the collection of samples (livers and raw meat) from pigs at participating Physi-Trace export processors, and represented two large, two medium and one small grower from each processor. Seven samples were taken from each grower totaling 203 raw meat and 203 liver samples. The samples were digested in a mixed acid solution at 90°C overnight before dilution with high purity (18 MΩ) water and were analysed using Inductively Coupled Plasma Mass Spectrometry (ICP-MS) and Inductively Coupled Plasma Atomic Emission Spectroscopy (ICP-AES) to determine trace element profiles. Due to the accumulation of some elements in the livers, a reduced set of analytes (Table 1) was developed for accuracy.
|
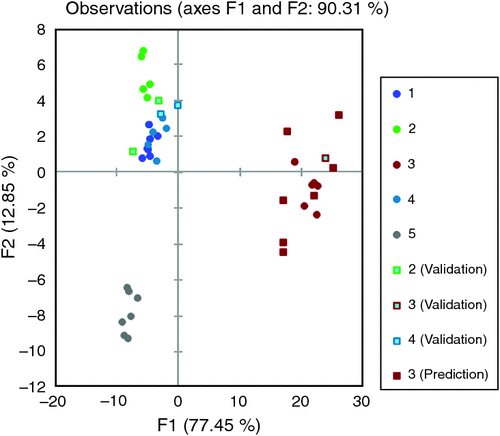
This investigation produced an algorithm that could be used to transform Physi-Trace elemental data for pork liver into ‘equivalent’ data that can be compared with pork meat data to determine property of origin. Fig. 1 is a linear discriminant analysis plot of the raw meat elemental data from all five growers from a specific processor with the addition of the transformed liver data from Grower 3.
|
From Fig. 1, all five growers can be easily discriminated and the transformed liver data for Grower 3 groups well with the raw meat data from Grower 3. Similar results were obtained for 86% of all the liver samples tested showing that the developed algorithm could be used to transform pork liver elemental data into data that could be used to compare with the elemental data of raw meat in the Physi-Trace database to assess the origin of the liver sample. To trace back to a processor of origin, the success rate of the algorithm was 97%.
References
Kreitals N (2013) Use of multi-element profiling for the traceability of Australian pork offal and its relationship to the pork meat physi-trace database. PhD Thesis.Supported by Australian Pork Limited.


