Estimating Relative Disulfide Energies: An Accurate Ab Initio Potential Energy Surface
Naomi L. Haworth A , Jason Y. Liu A , Samuel W. Fan A B , Jill E. Gready C and Merridee A. Wouters A B DA Structural & Computational Biology Division, Victor Chang Cardiac Research Institute, Sydney, NSW 2010, Australia.
B School of Medical Sciences, University of New South Wales, Sydney, NSW 2052, Australia.
C John Curtin School of Medical Research, Canberra City, ACT 2601, Australia.
D Corresponding author. Email: m.wouters@victorchang.edu.au
Australian Journal of Chemistry 63(3) 379-387 https://doi.org/10.1071/CH09456
Submitted: 31 August 2009 Accepted: 20 November 2009 Published: 26 March 2010
Abstract
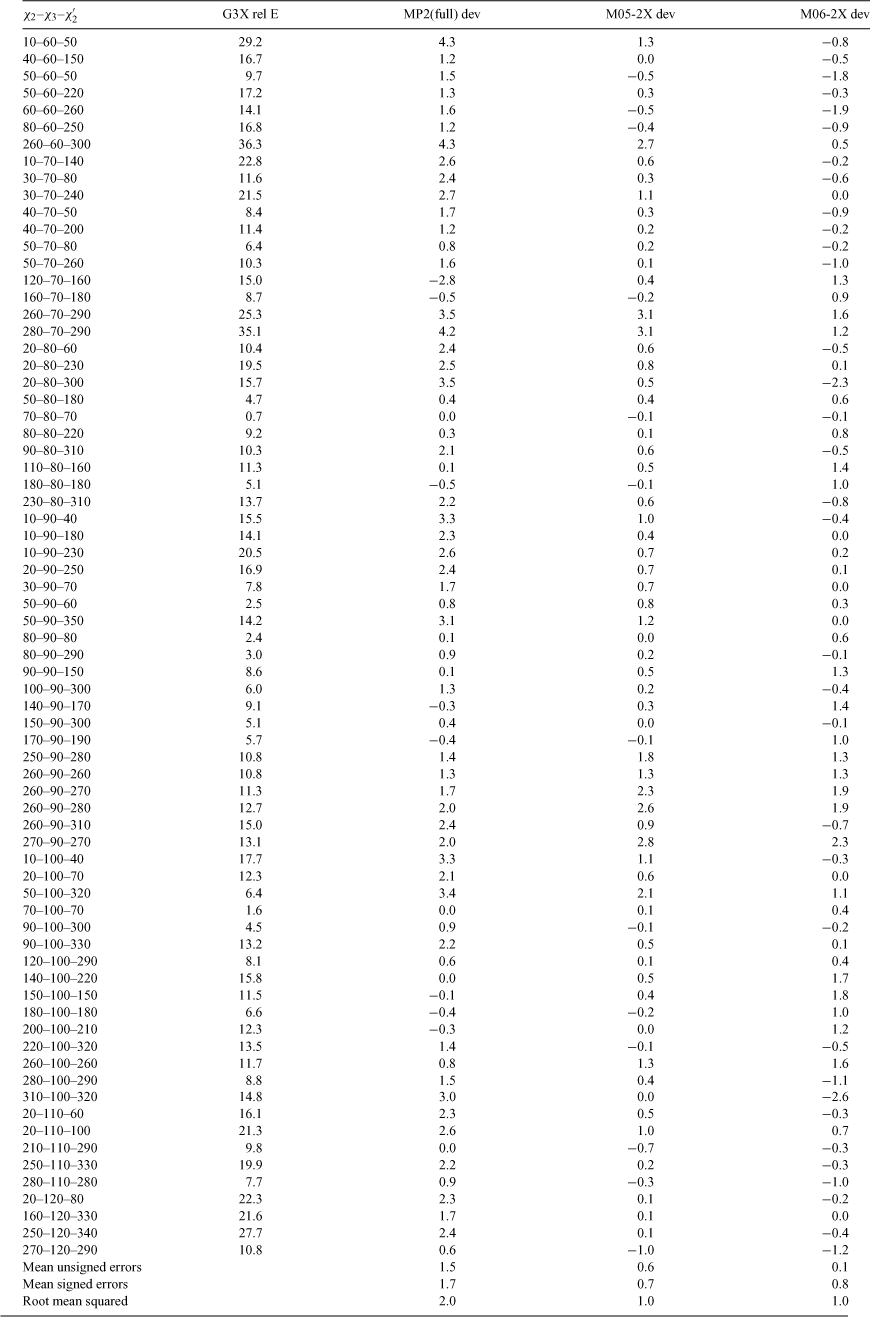
Disulfide torsional energy, a good predictor of disulfide redox potential in proteins, may be estimated by interpolation on a potential energy surface (PES) describing the twisting of diethyl disulfide through its three central dihedral angles. Here we update PES calculations at the M05-2X level of theory with the 6-31G(d) basis set. Although the surface shows no qualitative differences from an earlier MP2(full) PES, energy differences greater than 1 kJ mol–1 were seen for conformations with χ2 between –60° and 30°, or with χ3 below 60° or above 130°. This is particularly significant for highly strained disulfides that are likely to be spontaneously reduced by mechanical means. In benchmarking against the high-level G3X method, M05-2X showed significantly reduced root mean squared deviation compared with MP2(full) (1.0 versus 2.0 kJ mol–1 respectively). Results are incorporated into a web application that calculates relative torsional energies from disulfide dihedral angles (http://www.sbinf.org/applications/pes.html).
Introduction
Disulfides are conventionally viewed as structurally stabilizing elements in proteins but emerging evidence suggests two disulfide subproteomes exist. One group mediates the well known role of structural stabilization. The second redox-active group is best known for its catalytic functions but is increasingly being recognized for roles in the regulation of protein function. Disturbance of thiol-based redox regulation is implicated in diseases of oxidative stress such as neurodegenerative disease, cardiovascular disease, type II diabetes and cancer, as well as the aging process. An understanding of molecular processes underpinning these diseases will enable the development of diagnostics and therapeutics. Computational methods for distinguishing redox-active and structural disulfides in protein structures would be a boon.
The ultimate arbiter of protein disulfide redox activity is the redox potential of the disulfide relative to the redox potential of its milieu such as the biological compartment in which the protein resides. Disulfide redox potentials measured in thiol-disulfide oxidoreductases range from –95 to –330 mV.[1–4] For structural disulfides, the redox potential can be as low as –470 mV.[5] Disulfide redox potential is a function of electrostatic screening, entropic effects, and physical stress applied to the disulfide bond both through stretching and twisting. The redox potential contains both pH-dependent and pH-independent parts.[6] Thus pKa, which is often used to estimate redox potential, is not entirely predictive. As a practical matter, the calculation of pKa is not possible in a high-throughput manner.
Disulfide torsional energy is one variable that shows good correlation with the redox potential. Torsional strain on the disulfide bond is imposed by geometric constraints of the protein fold. Disulfide bonds are most stable when the CβSγSγ′Cβ′ dihedral angle adopts values close to 90°. Each sulfur atom has a lone pair of electrons in a p orbital. When the dihedral angle is 90° these lone pairs are orthogonal to each other and do not interact. Torsion of the disulfide bond, by either compression towards 0° or extension towards 180°, forces the orbitals to overlap, resulting in lone pair–lone pair repulsion which destabilizes the system. The result is a longer and weaker disulfide bond that is more susceptible to redox attack.[7,8]
Whitesides and co-workers used molecular mechanics calculations to investigate factors affecting redox potential of two thiols in molecules with similar structure and thiol pKa.[9] The most important parameters identified were the three central dihedral angles of the disulfide bond (χ2, χ3, and χ2′). Experimental work has confirmed torsional energy is an important variable influencing disulfide redox activity.[10–12] An empirical function from the AMBER force-field[13] was previously used to estimate disulfide torsional energies in a high-throughput manner using solved protein structures.[14] While this function has the right general form, it clearly does not take into account steric interactions within the system. A better description of the relative stabilities which includes steric effects is obtained by extending the AMBER analysis to include non-bonded terms, but this also gives inaccurate results.[15]
Quantum chemical calculations of torsional energy for the model compound diethyl disulfide were first attempted by Görbitz.[16] The diethyl disulfide model system is too small to allow investigation of the χ1 and χ1′ dihedral angles, however, it is possible to determine how the energy of the system changes as χ2, χ2′, and χ3 are varied. This is where the majority of the inaccuracy in the AMBER function is expected to lie. Görbitz found it necessary to use MP2 theory[17,18] with full correlation of core electrons in order to obtain an adequate description of the system. He used this method to identify salient features of the torsional energy surface such as the positions of extrema and inflexion points.
Building on Görbitz’ work, we used MP2(full) with the 6-31G(d) basis set to generate a full potential energy surface (PES) for torsion of diethyl disulfide around its three critical dihedral angles (χ2, χ3, and χ2′).[15] The use of 10 degree increments for each of the parameters resulted in a continuous, fine-grained surface. The MP2(full) surface showed significant qualitative differences from the PES calculated using the AMBER force-field. For instance, the AMBER PES predicts all local minima to be equally stable, whereas the MP2(full) surface shows more variation. Of particular importance is a high energy region that is completely unpredicted by the AMBER function. We showed that a reasonably flat ledge on this high energy feature was unexpectedly populated by disulfides which adopt the eclipsed or ‘staple’ conformation.[19] Optimized geometries for this conformation revealed that the χ3 dihedral angle is stretched to ~110°[15] with a corresponding lengthening of the SS bond.[20] The majority of these disulfides are found in a cross-strand context, that is, they covalently link Cys residues on adjacent strands of a β-sheet.[21] Backbone hydrogen bonds of the surrounding β-sheet help stabilize the high torsional energy disulfide conformation. Many of these ‘cross-strand disulfides’ have been associated with functional redox activity.[14,22,23] Interpolation on the MP2(full) PES[15] allowed us to predict the relative stabilities of disulfide bonds in high resolution structures from the Protein Data Bank (PDB).[24]
Prior to our earlier work there were relatively few computational investigations into the torsion of model compounds that emulated protein disulfide bonds. Earlier investigations generally focussed on smaller compounds, particularly disulfane and its halogenated analogues, which may be of interest to atmospheric scientists.[7,8,25–29] As shown by Aquilanti,[29] the highly electronegative halogen substituents significantly affect the nature of the disulfide bond, limiting the application of these studies to protein systems. Several groups have since reported investigations into alkyl disulfide torsion.[20,29,30] Of particular interest to proteins are studies by Dumont[30] and Galant[20] which investigated the effects of torsion on the electron affinity and dissociation energy of the disulfide bond, respectively.
During our original investigation we also explored the possibility of using density functional theory (DFT) to generate the PES. This would have had the advantage of significantly reducing the cost of calculations and providing the opportunity for further investigations on larger model systems. Unfortunately benchmarking calculations revealed that the best recognized DFT method of the time, B3LYP,[31] gave an inadequate description of the system. This is consistent with the findings of other workers using a variety of DFT methods.[30,32] Recently, new density functional methods have been introduced by Truhlar. The development of these methods, known as M05[33] and M05-2X,[34] has focussed on improving the way the kinetic-energy density is incorporated in exchange and correlation functionals, and removing self-correlation effects. Of particular interest to us is the M05-2X method, which incorporates twice the amount of Hartree–Fock exchange (hence 2X) in order to give a superior description of systems without metals. More recently, Truhlar introduced further improvements to these methods with updated functionals labelled M06 and M06-2X.[35,36] Here we investigate the use of the M0n functionals for describing the torsion of diethyl disulfide and recalculate the PES at the M05-2X level of theory. We examine the differences between the MP2(full), M05-2X, and M06-2X surfaces, particularly with regard to regions of the surface relevant for disulfides found in protein crystal structures. We have also implemented a web application that uses the new M05-2X PES to calculate relative disulfide torsional energies in protein structures using the five dihedral angles as input. The web application enables researchers in thiol-based redox biology to identify potential redox-active disulfides in X-ray crystal structures based on torsional energy.
Results
Benchmark Calculations
Results of the benchmarking calculations are given in Table 1. For each datum point, deviation of the relative energy from the benchmark G3X(CC) result is shown for each of the three methods of interest: MP2(full), M05-2X, and M06-2X, respectively. Mean unsigned errors (MUE), mean signed errors (MSE), and root mean squared (RMS) deviations are reported for each method. For almost all data points, the new M0n density functional methods give superior agreement with benchmarks in comparison with pure ab initio MP2(full) theory. Overall the MSE for MP2(full) is roughly two and a half times that of the M0n methods and the RMS deviation is twice as large. This is consistent with other reports that M05-2X can be superior to MP2 theory for describing relative isomer energies and non-covalent interactions.[37,38] On average M05-2X and M06-2X perform equally well, although for some data points, particularly those where χ2 and χ2′ are ~280° and χ3 < 80°, M06-2X is clearly much better. In other regions, however, most importantly where χ2 and χ2′ are ~180°, the older M05-2X methods give results closer to the benchmarks. In order to determine which of these two methods is preferable overall it was necessary to calculate the full PES and compare the performance of the methods for conformations adopted by disulfides found in X-ray crystal structures of the PDB.[24]
Potential Energy Surfaces
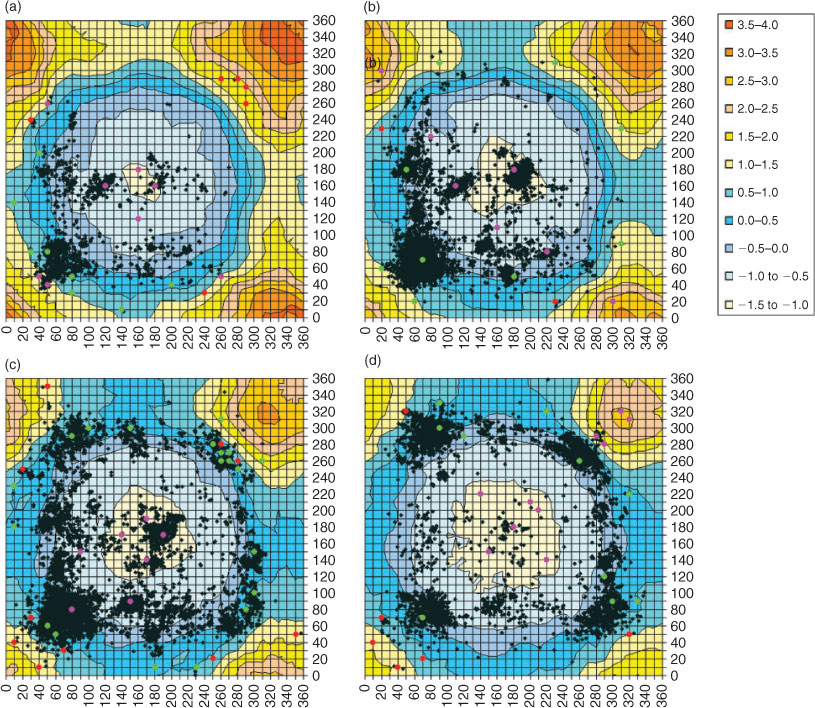
A full PES was calculated at the M05-2X/6-31G(d) level of theory. As it did not show significant qualitative differences from the MP2(full) surface reported in our previous paper, it is not reproduced here. Contour plots from biologically relevant slices through the PES (60° ≤ χ3 ≤ 120°) can be found in Appendix 1 (see Accessory Publication). Of greater interest are differences between the M05-2X and MP2(full) surfaces. Heat maps of these differences are shown in Fig. 1 for 60° ≤ χ3 ≤ 120°. Blue areas represent regions of the PES where the two methods differ by less than 1 kJ mol–1, whereas yellow to red regions show increasing deviations. MP2(full) calculations generally predict a higher relative torsional energy than M05-2X, as seen in the red, yellow, and greenish-blue regions of the heat map. For negative values in purple-blue areas of the figure, the reverse is true. Clearly over most of the difference plot the two surfaces do not differ greatly, however, as χ2 or χ2′ approaches 0°, MP2(full) predicts a much greater energy rise than M05-2X. The same effect is seen when χ3 falls below 60°. Benchmarking calculations confirm the M05-2X results, indicating MP2(full) is overpredicting destabilization of the system because of steric interactions. However, very few disulfides in proteins have χ2 or χ2′ close to 0° or χ3 below 60°. For practical purposes, therefore, we focussed on finding a surface accurate for regions of the PES where protein disulfides are found. We have, therefore, superimposed the conformations of all disulfides in X-ray crystal structures of the PDB which have χ3 within 5° of the value for the surface over each plot. This immediately reveals there are some regions of the PES where differences between the MP2(full) and M05-2X surfaces have an appreciable impact on our prediction of the relative torsional strain of a disulfide. These include the low energy spiral conformation (χ2 ~60°, χ2′ ~60°) for χ3 below 80° – lower left hand side (LHS) of Fig. 1a and 1b; the hook conformation (χ2 ~60°, χ2′ ~300°) for χ3 = 80° or 90° – lower right hand side (RHS) and upper LHS of Fig. 1c and 1d; and the high energy staple conformation (χ2 ~280°, χ2′ ~280°) for χ3 = 120° – upper RHS Fig. 1g and some examples with lower χ3. Differences between the surfaces in the staple region are particularly important because the torsional strain seen in staple disulfides is believed to be sufficiently high to destabilize the bond, rendering it susceptible to redox activity.
In early stages of this work, the M06-2X method was not yet available in the Gaussian program suite and thus could not be used in automatic generation of a PES. With the release of Gaussian 09, this has become possible and thus the M06-2X PES has been calculated for χ3 between 70° and 100°. Contour plots of this section of the PES can be found in Appendix 2 (see Accessory Publication). This has allowed a full comparison between the M05-2X and M06-2X functionals. Plots of the differences between the two methods (M05-2X and M06-2X) are shown in Fig. 2. As expected, deviations are much smaller than seen between MP2(full) and M05-2X. There are, however, some regions where the two methods differ by more than 1 kJ mol–1. In particular, M05-2X predicts higher destabilization in the region around χ2 ~300° and χ2′ ~300°, and M06-2X predicts the most extended conformations (χ2 ~180° and χ2′ ~180°) to be less stable than indicated by M05-2X.
In order to determine which method is most suitable for our purposes, conformations of disulfides in protein structures deposited in the PDB were superimposed on the PES slices (Fig. 2). The locations of the benchmarking points are also included. These are colour coded so that red points indicate benchmark conformations where the M06-2X result was more than 0.5 kJ mol–1 closer to G3X(CC) than M05-2X, pink points show where M05-2X was more than 0.5 kJ mol–1 better, and green points show where the two methods are equivalent. We note that all benchmarking points have been shown, including those not associated with disulfide populations. The latter were calculated to investigate differences between the MP2(full) and M05-2X surfaces but are unlikely to be of practical relevance for protein disulfides. Fig. 2 shows that for most of the benchmark results that coincide with significant disulfide populations in proteins, both M0n methods perform equally well. For our purposes, however, it is significant that for points through the centre of the plots (χ2 ~180° and χ2′ ~180°, where M05-2X predicts higher stability than M06-2X) the benchmarks indicate the M05-2X results are preferable. Also worth mentioning are the pink points in the region of χ2 ~280°, χ2′ ~280° for χ3 = 100°. Although a significant difference (>2.5 kJ mol–1) is seen here between the two methods, the region is not populated by protein disulfides. One might be misled into assuming this is a high energy region of the PES based on the difference plots, however, comparison with PES in the appendices reveals this region is a minimum with energy below 10 kJ mol–1, and thus is energetically accessible. This region may not be populated by disulfides in protein structures because of other conformational considerations. Table 1 shows that for this region M05-2X is in very good agreement with the benchmark, while M06-2X predicts disulfides in this region to be much more stable than they really are. There are also regions, however, where the M06-2X results are significantly better than M05-2X. These are most commonly regions where the relative energy is over 20 kJ mol–1, indicating that M06-2X better describes steric repulsion than M05-2X. There are, however, also some low energy points, such as χ2 = 30°, χ2′ = 70°, and χ3 = 90°, where the energy is less than 10 kJ mol–1 and M06-2X outperforms the older method.
The region around χ2 = 270°, χ2′ = 270°, and χ3 = 90° is of particular interest to us because it corresponds to the cross-strand disulfide secondary structural motif.[14,22] This motif was originally predicted to be forbidden, or disallowed, in protein structures because formation of the disulfide bond introduces strain in the resident protein structure due to the opposing stereochemical requirements of the disulfide bond and the resident β-sheet secondary structure.[39,40] Subsequent studies on increasingly larger populations of protein structures available in later releases of the PDB revealed disulfides are found in these ‘disallowed’ secondary structure contexts and seem to utilize the energetics of their situation to adopt a functional redox role in their resident proteins.[39,40] Because of its biological significance, accuracy in this region is particularly important to us, hence the high number of benchmark points. Here M06-2X is roughly 0.5 kJ mol–1 better than M05-2X (hence some points are green and others red), however, Table 1 shows that this difference is in the same direction and that M05-2X predicts the energy at this point to be nearly 3 kJ mol–1 too high. Comparison with neighbouring points indicates this relative deficiency in M05-2X is over a very small region – unfortunately it happens to be one of particular interest. Nevertheless, it is not possible to chop and change between methods in creating a PES. In general, for conformations adopted by most disulfides in the PDB, M05-2X seems to be equivalent or superior to M06-2X and thus this is the surface we have used in our web application. Where more accurate information is needed for cross strand disulfides, benchmark results are available in Table 1.
Torsional Energy Calculator
The torsional energy calculator is available at http://www.sbinf.org/applications/pes.html. Five dihedral angles are required as input. A screen shot of the application is shown in Fig. 3. Dihedral angles, entered in degrees, can be measured for input using standard protein visualization programs such as PyMol (DeLano Scientific LLC), SPDBV,[41] and Discovery Studio Visualizer (Accelrys). An interactive diagram of a disulfide helps users identify atoms that specify each torsional angle. All results are absolute energies relative to the lowest minimum on the surface. Zero-point energies are not included as calculation of these at every point of the three-dimensional PES is too computationally expensive. Results should be regarded as having an uncertainty of ±1.0 kJ mol–1 unless otherwise stated.

|
The use of NMR structures for the determination of disulfide torsional energies is not recommended. NMR structures are commonly determined using the magnetic dipole moment of hydrogens within the polypeptide chain. Because the common isotope of sulfur does not have a magnetic dipole moment, positions of sulfur atoms within a disulfide are inferred by triangulation of nearby hydrogens and are, therefore, not accurate.
Discussion
The PES is a valuable tool for more accurately estimating disulfide torsional energy. Protein disulfides occupying minima are likely to be structural disulfides while those in higher energy regions are likely to be redox active. However, a low disulfide torsional energy does not absolutely preclude redox activity of a disulfide. Disulfides in lower energy regions may become redox active if twisted into a higher energy conformation by a protein conformational change.[42] For cases such as these, inspection of other features of the protein in the vicinity of the disulfide with standard structural bioinformatic tools should give a clue as to whether the region is under stress. For any particular disulfide conformation, the redox potential of a disulfide is also influenced by other factors including electrostatic effects of residues nearby in space, and entropic effects associated with the polypeptide chain.
Disulfides with very high torsional energies (~17.5 kJ mol–1 or higher) are likely to be redox active and may be reduced by purely mechanical means such as stretching and twisting. An example is a short-lived disulfide intermediate formed between Cys 10 and Cys 82 as part of a disulfide cascade during the reaction cycle of S. aureus arsenate reductase (ArsC).[43] Our new PES predicts this disulfide to have a relative torsional energy of 18.5 kJ mol–1.
In addition to very unstable high torsional energy disulfides, other disulfides of intermediate energy (10–17.5 kJ mol–1) are also likely to be redox active. The majority of ‘forbidden’ disulfides, a set of canonical disulfide motifs in primary and secondary structure with non-optimal stereochemistry,[22,23] fall into this group. Forbidden disulfides account for nearly 30% of intermediate energy disulfides, but constitute only 9% of disulfides with torsional energies below 10 kJ mol–1 and 23% of those over 17.5 kJ mol–1. Their metastability is likely an important feature of their biological function. While most forbidden disulfides are unlikely to undergo spontaneous reduction, they are likely to be far more susceptible to redox attack than low energy, structural disulfides. Their reduction may need to be assisted by an oxidoreductase, such as thioredoxin.
Torsional energies may be useful discriminators of the likely behaviour of particular forbidden disulfides. Examples of the recently identified canonical forbidden disulfide known as the end-twist[23] are illustrative. The previously mentioned high-torsional energy disulfide in ArsC is an example of an end-twist disulfide, which is formed as part of a disulfide cascade during the ArsC reaction cycle which detoxifies arsenic compounds. Other examples of end-twist disulfides are found in the dual-specificity phosphatases PTEN and PRL-1, which are homologous to ArsC but have different biological functions. Dual specificity phosphatases dephosphorylate Thr, Ser, and Tyr residues, as well as inositol phospholipids, as part of intracellular signalling pathways.[44,45] Oxidation inactivates PTEN and PRL-1 by formation of an end-twist disulfide between the catalytic Cys and another nearby Cys. The inhibitory end-twist disulfide which forms in PRL-1 between Cys 49 and Cys 104 has a relative torsional energy of 5.3 kJ mol–1 (PDB: 1zck), which is quite stable for a forbidden disulfide. Some biochemical evidence suggests PTEN may be reduced by thioredoxin, which is consistent with the lower torsional energy of the homologous disulfide in PRL-1, which would need a strong reductant, and possibly conformational priming, to reduce it. Thus, although ArsC and the dual specificity phosphatases both harbour disulfides in end-twist motifs, their torsional energies suggest different stabilities, and hence different modes of reduction, which are consistent with current biochemical evidence. This also makes sense from the perspective of timeframe of biological function, as the mechanically reduced higher torsional energy end-twist motif in ArsC is part of a reaction cycle, whereas the lower energy end-twist in PRL-1, which is likely dependent on an exogeneous protein for reduction, is an inhibitory disulfide in a signalling pathway.[46] Thus the expected biological lifetimes of the disulfides are consistent with the calculated relative torsional energies.
We note that disulfides with a predicted relative torsional energy above 25 kJ mol–1, and particularly those with energies over 30 kJ mol–1, should be regarded with a degree of caution. While it is possible that the disulfide really is so highly strained in the biological system, there is also a significant risk that the conformation is an artifact of poor modelling or very low resolution X-ray data.
Conclusion
In summary, a M05-2X quantum chemical potential energy surface has been generated to improve estimates of disulfide torsional energies. The calculations have been used as the basis of a web application which should be of value in judging the likelihood of disulfide redox activity. Very high (≥17.5 kJ mol–1) and high (10–17.5 kJ mol–1) torsional energy disulfides are likely to be redox active. Low torsional energies do not preclude disulfide redox activity as other factors aside from torsional energy may be relevant. The web application should aid researchers investigating the molecular basis of thiol-based redox biology.
Experimental
Potential Energy Calculations
As in our previous work, a PES was generated for torsion around the three central dihedral angles (χ2, χ3, and χ2′) of the diethyl disulfide model compound. Full geometry optimizations were performed at the M05-2X/6-31G(d) level of theory for each 10 degree increment of each of the three variables. Energies are reported relative to the fully optimized global minimum of the surface (χ2 ~68°, χ3 ~87°, χ2′ ~68° in all cases). A similar PES was calculated at the M06-2X/6-31G(d) level of theory for χ3 between 70° and 100°. Extensive benchmarking calculations were performed at a variety of points scattered across the PES. These points focussed on regions of the PES where differences of more than 1 kJ mol–1 were seen either between the M05-2X relative energies and those predicted by our previous MP2(full)/6-31G(d) surface or between the M05-2X and M06-2X results. Points were also chosen to ensure that regions of the PES most highly populated by disulfides in the PDB were represented. Benchmark calculations were performed using the high-level G3X(CC) ab initio theory with B3LYP/6-31G(2df,p) reference geometries. Zero-point energies were not included in the G3X results to give better comparison with relative energies calculated at the lower levels of theory. G3X and M05-2X calculations were performed using the Gaussian 03 suite of programs.[47] Initial M06-2X calculations were performed using Molpro,[48] with the Gaussian 09 suite[49] being used for later, more extensive testing of M06-2X. Calculations were performed on the AC and XE computer clusters of the Australian Partnership of Advanced Computing (APAC) National Facility.
Torsional Energy Calculator
A web application was developed to enable easy calculation of disulfide torsional energies in protein structures by redox biologists. M05-2X PES data are held in a postgres SQL database. An html interface was built that executes a php script after the user enters the five disulfide dihedral angles and hits the submit button. Within the php script, simple checks are made at the input stage to ensure dihedral angles are within bounds. To calculate the energy contributions of the three central dihedral angles χ2, χ3, and χ2′, the eight grid points of the cube surrounding the point of interest are retrieved from the SQL database and a linear interpolation is performed between them. Contributions due to χ1 and χ1′ of the disulfide are incorporated using the relevant terms from the AMBER function such as 2(2 + cos 3χ1 + cos 3χ1′) (kcal mol–1). The relative torsional energy is output in kJ mol–1.
Accessory Publication
Contour plots of slices through the M05-2X/6-31G(d) and M06-2X/6-31G(d) 3D-PES for diethyl disulfide are available on the Journal’s website.
Acknowledgements
This research was undertaken on the NCI National Facility in Canberra, Australia, which is supported by the Australian Commonwealth Government. The authors would like to thank Dr Siiri Iismaa for critical comments on the manuscript.
[1]
M. Wunderlich,
R. Glockshuber,
Protein Sci. 1993, 2, 717.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
|
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
|
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
|
CAS |
| Crossref | GoogleScholarGoogle Scholar |
| Crossref | GoogleScholarGoogle Scholar |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |
| Crossref | GoogleScholarGoogle Scholar |
CAS |

[47]
[48]
[49]