The Influence of Secondary Structure on Electron Transfer in Peptides
Jingxian Yu A B , John R. Horsley A and Andrew D. Abell A BA School of Chemistry and Physics, The University of Adelaide, Adelaide, SA 5005, Australia.
B Corresponding authors. Email: jingxian.yu@adelaide.edu.au; andrew.abell@adelaide.edu.au
Australian Journal of Chemistry 66(8) 848-851 https://doi.org/10.1071/CH13276
Submitted: 30 May 2013 Accepted: 16 July 2013 Published: 29 July 2013
Abstract
A series of synthetic peptides containing 0–5 α-aminoisobutyric acid (Aib) residues and a C-terminal redox-active ferrocene was synthesised and their conformations defined by NMR and circular dichroism. Each peptide was separately attached to an electrode for subsequent electrochemical analysis in order to investigate the effect of peptide chain length (distance dependence) and secondary structure on the mechanism of intramolecular electron transfer. While the shorter peptides (0–2 residues) do not adopt a well defined secondary structure, the longer peptides (3–5 residues) adopt a helical conformation, with associated intramolecular hydrogen bonding. The electrochemical results on these peptides clearly revealed a transition in the mechanism of intramolecular electron transfer on transitioning from the ill-defined shorter peptides to the longer helical peptides. The helical structures undergo electron transfer via a hopping mechanism, while the shorter ill-defined structures proceeded via an electron superexchange mechanism. Computational studies on two β-peptides PCB-(β3Val-β3Ala-β3Leu)n–NHC(CH3)2OOtBu (n = 1 and 2; PCB = p-cyanobenzamide) were consistent with these observations, where the n = 2 peptide adopts a helical conformation and the n = 1 peptide an ill-defined structure. These combined studies suggest that the mechanism of electron transfer is defined by the extent of secondary structure, rather than merely chain length as is commonly accepted.
Introduction
Electron transfer occurs in proteins over surprisingly long molecular distances of up to 100 Å to facilitate, for example, the function of mitochondria and the process of photosynthesis in green plants and algae.[1,2] Several factors are thought to influence the kinetics of these processes, including peptide chain length, dipole orientation, backbone conformation, and hydrogen bonding and hence secondary structure.[3] However, the exact role that these and other factors play in defining the associated mechanisms remains somewhat contentious.[4] A fundamental understanding of how electron transfer occurs in proteins is needed to progress the field and also to provide an opportunity for the development of applications in the area of molecular electronics.[5,6] With this in mind, we recently initiated experimental and theoretical studies on electron transfer in a series of model peptides that are known to adopt secondary structure.
Mechanistic Transition
Two mechanisms, namely bridge-assisted superexchange and electron-hopping, were originally proposed to rationalise long-range electron transfer in DNA, particularly the effect of the separation of the donor and acceptor (distance dependence) groups.[7] However, these mechanisms remain somewhat controversial, with conflicting reports on their application to peptides.[8,9] In an attempt to begin to clarify this we recently reported[10] electron transfer studies on a series of oligomers [H-(NH-C(CH3)2-CO)n-NH-CH2-Fc, n = 0–5] of α-aminoisobutyric acid (Aib) that contain a C-terminal ferrocene (Fc) for electrochemical studies and a free N-terminus for attachment to a single-walled carbon nanotube array/p-silicon (100) electrode. Oligomers of geminally disubstituted Aib units were used in this study since relatively short sequences (3 or more residues) are known to induce predictable and stable 310-helical structures.[11] The conformations of these peptides were confirmed by 2D NMR to be helical for the longer synthetic peptides (n = 3–5) and ill-defined for the shorter peptides (n = 0–2). The shorter peptides cannot adopt a 310-helix since the requisite i to i+3 intramolecular hydrogen bonds cannot form.
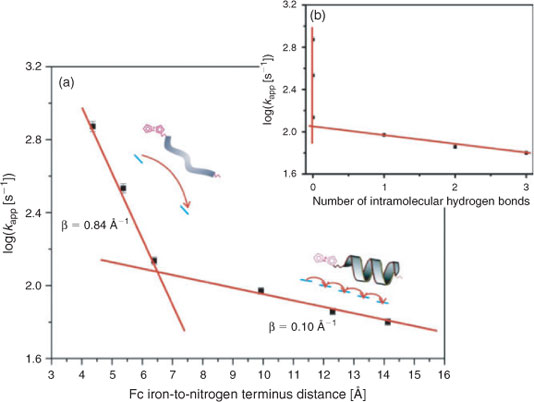
The electron transfer rate constants (kapp) were determined for each of the peptides and these values were plotted against the Fc iron-to-nitrogen terminus distance as shown in Fig. 1a. A slope transition is apparent for the peptide containing three Aib units (see the 4th data point from the left in this Figure).[10] It should be noted that the efficiency of electron transfer in peptides is measured by the attenuation constant (β), which provides information on the electron transfer mechanism.[12] Shorter sequences (n = 0–2) gave rise to a steep decrease on increasing the chain length. The apparent rate attenuation constant (β) for these peptides was calculated to be 0.84 Å–1.[10] This is consistent with an electron superexchange mechanism.[7] The longer helical peptides (n = 3–5) display a weak dependence of the electron transfer rate constant on distance. The rate attenuation constant (β) for these peptides was estimated to be 0.10 Å–1, as evidenced by the shallow slope of the last 3 points,[10] indicative of an electron hopping mechanism.[7]
|
The corresponding electron transfer rate constants (kapp) were plotted against the number of intramolecular hydrogen bonds as shown in Fig. 1b.[10] The lack of intramolecular hydrogen bonding and hence secondary structure associated with the shorter peptides (n = 0–2) (see the first three data points from Fig. 1a) gives rise to electron transfer via a superexchange mechanism. By comparison, the longer peptides (n = 3–5) have a helical structure as defined by a series of well defined intramolecular hydrogen bonds. Here a strong dependence between kapp and the number of intramolecular hydrogen bonds is observed. It is likely that intramolecular hydrogen bonding facilitates electron transfer in structurally well defined peptides by way of a hopping mechanism. This study suggests that the mechanism of electron transfer is dictated by the extent of secondary structure and hence the degree of intramolecular hydrogen bonding, rather than merely by chain length.
Electron Transfer Dynamics
Two β-peptides [PCB-(β3Val-β3Ala-β3Leu)n–NHC(CH3)2OOtBu, n = 1 and 2, designated β3 and β6 respectively; PCB = p-cyanobenzamide] were also synthesised and their electron transfer studied in order to gain further insight and support for our earlier observations.[13] β-Peptides were used in this study as they are known to adopt helical secondary structures with high predictability, structural diversity, and stability.[14–18] These structures are ideal for studying the fundamental mechanisms of electron transport, but they have as yet received scant attention. In this case, an N-terminal tert-butyl hydroperoxide was used as the electron acceptor and a C-terminal p-cyanobenzamide as the donor. By using hydroperoxide and cyanobenzamide, the electrochemistry can be determined in solution, whereby the conformational freedom of the peptide is increased.[4] Circular dichroism (CD) and 2D NMR analysis on β6 revealed that its backbone readily folds into the expected low-energy hydrogen-bonded 314-helical structure,[3,19] which is consistent with gas-phase geometry optimisation using the density functional theory (DFT) method. Gas-phase geometry optimisation on β3 suggested that it adopts an ill-defined conformation. Electron transfer rate constants of 2580 s–1 and 9.8 s–1 were determined in solution for β3 and β6, respectively.[13] Based on these values it is clear that the two peptides have different efficiency of electron transfer, suggesting a likely difference in mechanism. Based on this a detailed computational study was undertaken as discussed below.
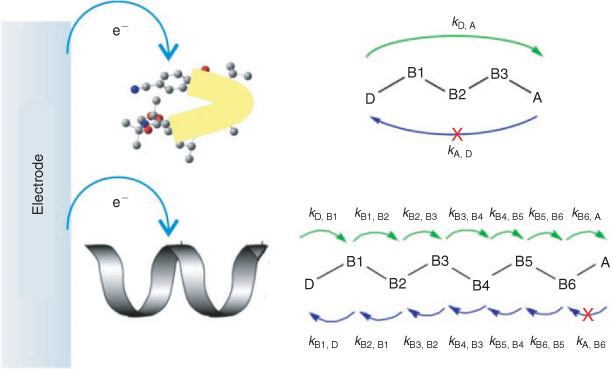
Latest constrained density functional theory (cDFT) calculations relating to the energy of each diabatic state for both β-peptides gave rise to accurate potential surfaces that were used to estimate the driving force (ΔG), reorganisation energy (λ), and the electronic coupling matrix element (Hab). These parameters allowed the electron transfer rate constant to be calculated using Marcus theory. The diabatic states in both β-peptides were obtained by individually localising an overall charge of –1 on each of the donor (D), amino acids, and the acceptor (A). Fig. 2 shows constructed diabatic states for both peptides and the possible electron transfer pathways.
For β3, the electron transfer rate constants for the forward steps D→B1, B3→A, and backward steps A→B3 along the peptide backbone are extremely small, indicating a highly improbable electron transfer route. This is consistent with a superexchange electron transfer pathway for this peptide as depicted in Fig. 3 (top). In comparison, the electron transfer rate constants for the forward (D→A) and backward (A→D) steps for β6 are exceptionally small (see Fig. 2). This implies that a superexchange mechanism does not occur in this case, with the alternative electron hopping pathway operating. These results provide theoretical evidence for the two model peptides following different electron-transfer pathways, presumably again due to different conformations and hence secondary structure.
|
Conclusion
Electron transfer studies on a series of oligomers of α-aminoisobutyric acids revealed a change from an electron superexchange to a hopping mechanism on transitioning from an ill-defined to a helical conformation. Further studies involving β–peptides provided theoretical evidence to corroborate these findings. Together these results clearly show that the mechanism of electron transfer in peptides is defined by the extent of secondary structure, rather than simply chain length. Other factors may influence the electron transfer mechanism in peptides, such as dipole orientation and the molecular dynamics of the backbone. Further research is pivotal to advancing the field and to provide opportunities for the use of peptides in molecular electronics applications. All this work is based on an ability to define and control the shape or conformation of peptides.[20–22]
References
[1] Y. Arikuma, H. Nakayama, T. Morita, S. Kimura, Langmuir 2011, 27, 1530.| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsVGktL%2FJ&md5=bf6f11887f0309fcc92983d59a152baeCAS | 21090665PubMed |
[2] G. P. Smestad, M. Gratzel, J. Chem. Educ. 1998, 75, 752.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXjtlajtb8%3D&md5=5ff7664051868f51edfc064ca4de0b74CAS |
[3] P. A. Brooksby, K. H. Anderson, A. J. Downard, A. D. Abell, Langmuir 2010, 26, 1334.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXht1WktLnO&md5=d02c9bdea561590bd0a12044f33824a4CAS | 19799404PubMed |
[4] Y. T. Long, E. Abu-Rhayem, H. B. Kraatz, Chem. – Eur. J. 2005, 11, 5186.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVCgsbzO&md5=d1367e59410527e9f4e405167afdee31CAS | 15977279PubMed |
[5] J. J. Davis, D. A. Morgan, C. L. Wrathmell, D. N. Axford, J. Zhao, N. Wang, J. Mater. Chem. 2005, 15, 2160.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXks1Cjsro%3D&md5=620e4842fd6460db22e1390efbbdcdbdCAS |
[6] Y.-S. Chen, M.-Y. Hong, G. S. Huang, Nat. Nanotechnol. 2012, 7, 197.
| Crossref | GoogleScholarGoogle Scholar | 22367097PubMed |
[7] B. Giese, J. Amaudrut, A. K. Kohler, M. Spormann, S. Wessely, Nature 2001, 412, 318.
| Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3MvhtF2isA%3D%3D&md5=75623591c92af6d959eb63042917b94fCAS | 11460159PubMed |
[8] R. A. Malak, Z. N. Gao, J. F. Wishart, S. S. Isied, J. Am. Chem. Soc. 2004, 126, 13888.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXot1Wns7k%3D&md5=eb2332a00e1f4f7ed4a9997800fb75b3CAS | 15506726PubMed |
[9] F. Polo, S. Antonello, F. Formaggio, C. Toniolo, F. Maran, J. Am. Chem. Soc. 2005, 127, 492.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtFCitbbO&md5=8d49985e90826b549f145e977e04e291CAS | 15643851PubMed |
[10] J. Yu, O. Zvarec, D. M. Huang, M. A. Bissett, D. B. Scanlon, J. G. Shapter, A. D. Abell, Chem. Commun. 2012, 48, 1132.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XivVOiug%3D%3D&md5=85330fdcf189b1ab87a69d7546de9519CAS |
[11] D. F. Kennedy, M. Crisma, C. Toniolo, D. Chapman, Biochemistry 1991, 30, 6541.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXkt1Ggs7g%3D&md5=07a02d14628fae9959b5889100b05d40CAS | 2054352PubMed |
[12] O. S. Wenger, Acc. Chem. Res. 2011, 44, 25.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXht12ju73F&md5=e153c12a2a6fedc473993b80ef59b9a5CAS | 20945886PubMed |
[13] J. Yu, D. M. Huang, J. G. Shapter, A. D. Abell, J. Phys. Chem. C 2012, 116, 26608.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsleqtbzJ&md5=721e949ec12f290d7e6f2c7f02e48743CAS |
[14] R. P. Cheng, S. H. Gellman, W. F. DeGrado, Chem. Rev. 2001, 101, 3219.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXntlGntbg%3D&md5=0e6dbd37f94caedfe78a857ab8d9c51aCAS | 11710070PubMed |
[15] D. S. Daniels, E. J. Petersson, J. X. Qiu, A. Schepartz, J. Am. Chem. Soc. 2007, 129, 1532.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXns1Ohsw%3D%3D&md5=4a3260686209b22a101881e1120712b1CAS | 17283998PubMed |
[16] C. M. Goodman, S. Choi, S. Shandler, W. F. DeGrado, Nat. Chem. Biol. 2007, 3, 252.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXkt1Kjs7w%3D&md5=3ed67326b8fa938d555d0f829ae7318fCAS | 17438550PubMed |
[17] J. A. Kritzer, J. Tirado-Rives, S. A. Hart, J. D. Lear, W. L. Jorgensen, A. Schepartz, J. Am. Chem. Soc. 2005, 127, 167.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVOjtrbJ&md5=43cc9e888a0ae3eaac1f6a0027ed0e19CAS | 15631466PubMed |
[18] D. Seebach, J. Gardiner, Acc. Chem. Res. 2008, 41, 1366.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXnvVKksrk%3D&md5=33c21c21a5475b2959af065f668fc118CAS | 18578513PubMed |
[19] P. A. Brooksby, K. H. Anderson, A. J. Downard, A. D. Abell, J. Phys. Chem. C 2011, 115, 7516.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjvFaqsLc%3D&md5=bfd9da47fb741b77116105d4e700540bCAS |
[20] A. D. Pehere, A. D. Abell, Org. Lett. 2012, 14, 1330.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XisVGht7g%3D&md5=b093f0eb2487d6b0aa7954c9d954d719CAS | 22339212PubMed |
[21] A. D. Abell, M. A. Jones, J. M. Coxon, J. D. Morton, S. G. Aitken, S. B. McNabb, H. Y. Y. Lee, J. M. Mehrtens, N. A. Alexander, B. G. Stuart, A. T. Neffe, R. Bickerstaff, Angew. Chem. Int. Ed. 2009, 48, 1455.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXisFCltrs%3D&md5=556aee4c73a91226dbaee5a4d0f3c370CAS |
[22] R. I. Mathad, B. Jaun, O. Floegel, J. Gardiner, M. Loeweneck, J. D. C. Codee, P. H. Seeberger, D. Seebach, M. K. Edmonds, F. H. M. Graichen, A. D. Abell, Helv. Chim. Acta 2007, 90, 2251.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFOqtLo%3D&md5=f8acef01612f8078471e57f9183c85aeCAS |



