Goldacre Paper:Understanding meiosis and the implications for crop improvement
Jason A. Able A C , Wayne Crismani A and Scott A. Boden A BA School of Agriculture, Food and Wine, The University of Adelaide, Waite Campus, PMB1, Glen Osmond, South Australia 5064, Australia.
B Present address: The John Innes Centre, Colney Lane, Norwich NR4 7UH, UK.
C Corresponding author. Email: jason.able@adelaide.edu.au
This paper originates from the Peter Goldacre Award 2008 of the Australian Society of Plant Scientists, received by the first author.
Functional Plant Biology 36(7) 575-588 https://doi.org/10.1071/FP09068
Submitted: 27 March 2009 Accepted: 1 May 2009 Published: 2 July 2009
Abstract
Over the past 50 years, the understanding of meiosis has aged like a fine bottle of wine: the complexity is developing but the wine itself is still young. While emphasis in the plant kingdom has been placed on the model diploids Arabidopsis (Arabidopsis thaliana L.) and rice (Orzya sativa L.), our research has mainly focussed on the polyploid, bread wheat (Triticum aestivum L.). Bread wheat is an important food source for nearly two-thirds of the world’s population. While creating new varieties can be achieved using existing or advanced breeding lines, we would also like to introduce beneficial traits from wild related species. However, expanding the use of non-adapted and wild germplasm in cereal breeding programs will depend on the ability to manipulate the cellular process of meiosis. Three important and tightly-regulated events that occur during early meiosis are chromosome pairing, synapsis and recombination. Which key genes control these events in meiosis (and how they do so) remains to be completely answered, particularly in crops such as wheat. Although the majority of published findings are from model organisms including yeast (Saccharomyces cerevisiae) and the nematode Caenorhabditis elegans, information from the plant kingdom has continued to grow in the past decade at a steady rate. It is with this new knowledge that we ask how meiosis will contribute to the future of cereal breeding. Indeed, how has it already shaped cereal breeding as we know it today?
Additional keywords: Asynapsis1, chromosome pairing, meiosis, Ph1, recombination, synaptonemal complex, wheat.
Introduction
Given the importance of small grain cereals including wheat (Triticum aestivum L.), rice (Oryza sativa L.) and barley (Hordeum vulgare L.) in agriculture, there is constant demand to produce new varieties. With world wheat stocks declining and rice yield reaching a plateau in recent years, it is imperative that demand is met for an ever expanding global population. Meeting this demand will have its challenges but through the marriage of classical plant breeding and molecular approaches to create new varieties, we have already moved forward significantly in the past two decades. Underpinning significant yield improvements in many of the cereal crops is the understanding of how cellular processes work and how those processes can then be manipulated for the benefit of plant breeding programs. One such process, that many scientists and plant breeders alike consider as the ‘holy grail’ in significantly being able to enhance plant breeding strategies of the future, is meiosis.
Meiosis is required for the production of gamete cells that contain half of the genome content of a parental cell. This halving of genome content ensures that, upon fertilisation, the newly formed zygote contains the correct amount of genetic information, equal in quantity to that of the parents’ cells. In addition, desirable combinations of alleles can be produced during meiosis. These combinations are the direct result of the recombination events that occur, ultimately leading to the genetic diversity that we see from generation to generation. However, strong selection pressure imposed through cereal breeding programs in the past has limited the genetic diversity that is readily available (Able et al. 2007).
Historically, meiotic studies in all eukaryotes have involved cytological analysis of cells by light and electron microscopy to understand key chromosomal events. Through such analysis the meiotic cycle has been divided into several stages, principally by the various changes in chromosome morphology that occur (Fig. 1). During one of the early stages, prophase I, there are a further five sub-stages (leptotene, zygotene, pachytene, diplotene and diakinesis), defined by cytological observations, during which three important meiotic events are occurring: chromosome pairing, synapsis and recombination (Table 1). These three events are intimately associated with each other. Following the alignment of homologous chromosomes during leptotene, the inter-homologue interactions of synaptonemal complex (SC) formation and recombination occur. While recombination is generally recognised to initiate before SC formation, it is thought that the SC stabilises interactions between homologous chromosomes to allow for resolution of recombination events into either crossovers or non-crossovers (reviewed in Kleckner 1996; Börner et al. 2004; Page and Hawley 2004). Recombination and SC formation have been the subject of extensive molecular analysis, and both have been implicated in the alignment and recognition of homologous chromosomes.
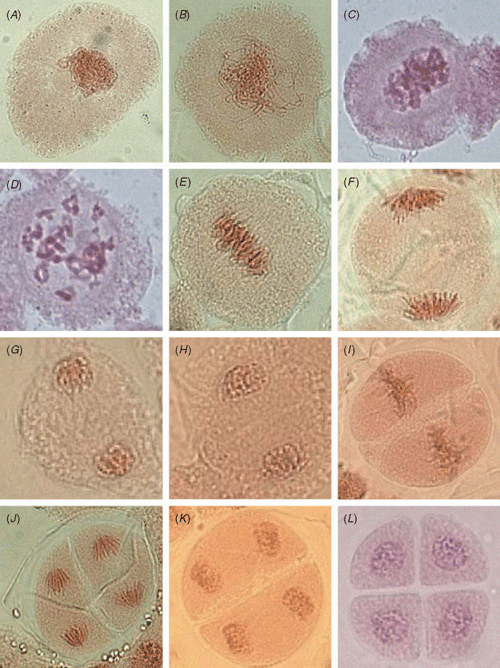
|

|
Model organisms such as budding yeast (Saccharomyces cerevisiae) have proven to be influential in understanding the factors and mechanisms regulating such key events during meiosis. Given that meiosis is an evolutionarily conserved process, many of the molecular events identified in budding yeast are also shared by higher eukaryotes. Even so, this has not necessarily meant that the amino acid sequence of proteins involved in such events is well conserved between yeast and higher eukaryotes; as the isolation of key meiotic genes and proteins in plants and animals based on sequence homology alone has sometimes been difficult. This has particularly been the case for genes that have a role in synapsis of homologous chromosomes (Caryl et al. 2000; Higgins et al. 2005). However, one excellent example of where conservation of function has been demonstrated in a wide variety of organisms is HOmolog Pairing 1 (HOP1) isolated from yeast (Hollingsworth and Byers 1989), otherwise known as ASYnapsis 1 (ASY1) in Arabidopsis (Arabidopsis thaliana L.) (Ross et al. 1997; Caryl et al. 2000), Brassica spp. (Armstrong et al. 2002), bread wheat (Boden et al. 2007) or homologous Pairing Aberration In Rice meiosis 2 (PAIR2) in rice (Nonomura et al. 2004). Identified as an integral component in not only chromosome pairing of yeast (Hollingsworth and Byers 1989) but now also bread wheat (Boden et al. 2009), this gene will be the focus of discussion later.
The key events during early meiosis
Homologous chromosome pairing
There are many different factors which contribute to chromosome homology recognition; including chromosome morphology, chromatin re-modelling, regions of DNA sequence homology, and proteins that interact with chromatin (Hamant et al. 2006). The juxtaposition and alignment of homologous chromosomes during meiosis is the least well understood process of prophase I, with the mechanism by which homologous chromosomes align still remaining somewhat unresolved. Contributing to this is the difficulty in establishing whether a loss of bivalent formation in a given mutant is the consequence of defective pairing or synapsis, as deficiency in both processes could equally yield univalents. Furthermore, various studies in fruit fly (Drosophila melanogaster) and the nematode Caenorhabditis elegans have clearly demonstrated that a uniform mechanism for homologous chromosome alignment does not exist (when compared with other model organisms) (Carpenter 1975; Goldstein and Slaton 1982; Orr-Weaver 1995; Fung et al. 1998; MacQueen et al. 2005).
Various stages of the recombination pathway have been suggested to contribute to homologous chromosome recognition. For example, studies in maize (Zea mays L.) have shown that there is a significant decrease in the number of RADiation sensitive 51 (RAD51) foci from the beginning of zygotene compared with pachytene, and the high numbers of zygotene foci have been proposed to support a role for RAD51 in homology searching (Franklin et al. 1999). This has been supported by observations that maize mutants with abnormalities in the distribution and numbers of RAD51 foci during prophase I also display defects in homologous chromosome pairing (Pawlowski et al. 2003, 2004). Similarly, the Arabidopsisasy1 mutant which fails to correctly synapse homologous chromosomes, also displays an abrupt decrease in Disrupted Meiotic cDNA 1 (DMC1) foci following its initial loading onto chromatin, relative to wild-type (Sanchez-Moran et al. 2007).
Another mechanism suggested to facilitate homologous chromosome alignment is the formation of a structure known as the telomere bouquet (reviewed in Zickler and Kleckner 1998; Harper et al. 2004). The telomere bouquet forms as telomeres of each chromosome attach to the nuclear periphery and cluster together during mid-prophase I (Hiraoka 1952; Zickler and Kleckner 1998). Formation of the telomere bouquet may assist homologous chromosome pairing by bringing the chromosomes into close association with one another, thereby decreasing the distance between homologous chromosomes (Moses 1968; Hamant et al. 2006). Studies in maize support this theory as the pam1 (plural abnormalities in meiosis 1) mutant, which displayed a loss of telomere bouquet formation, was also found to have a dramatic reduction in homologous chromosome pairing (Golubovskaya et al. 2002). However, there is also evidence to show that the telomere bouquet is not an essential requirement for homologous chromosome pairing, and that in several cases initial homologue interactions precede telomere bouquet formation (reviewed in Zickler and Kleckner 1998; Hamant et al. 2006).
Synaptonemal complex formation
The completion of homologous chromosome alignment and pairing is observed cytologically by the formation of a structure known as the SC. The SC is a tripartite protein structure that forms between non-sister chromatids of homologous chromosomes (von Wettstein et al. 1984; Fig. 2; also see Page and Hawley 2004). While its direct role in homologous chromosome interactions has been difficult to dissect, the evolutionary conservation of the structure across a wide range of species indicates that the SC does have a conserved role during meiosis (reviewed in Kleckner 1996; Hunter 2003).
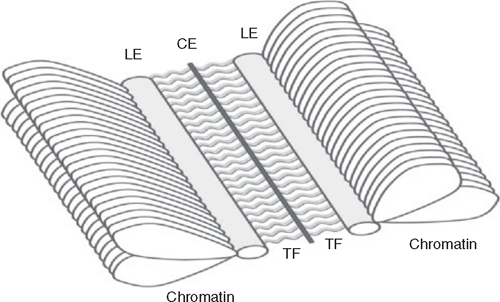
|
In association with chromosome condensation that occurs through leptotene, sister chromatids become organised along structures known as axial elements. During the early stages of synapsis, the axial elements mature into lateral elements (Rockmill et al. 1995). The lateral elements of non-sister chromatids are linked through recombination-mediated induced double strand breaks (DSBs), and become joined through the recruitment and formation of transverse filaments (reviewed in Page and Hawley 2004). In yeast, correct localisation of transverse filament proteins requires the presence of proteins involved in chromosome condensation, indicating a coordination of chromosome synapsis with the process of chromosome condensation (Klein et al. 1999; Yu and Koshland 2003). At pachytene, when homologous chromosomes have completely synapsed, the cell contains a complete SC complement (Albini and Jones 1988). The complete SC includes the lateral elements and transverse filaments, as well as a dense region, at equidistance from each lateral element, known as the central element (reviewed in Page and Hawley 2004). Entering diplotene, the SC begins disassembly as the cell progresses towards metaphase and anaphase, where chiasmata form the sole physical union between non-sister chromatids.
Recent evidence from studies in yeast, mammals and Arabidopsis indicate that formation of the SC and progression of the recombination pathway is intimately related (reviewed in Kleckner 2006). Studies in yeast and Arabidopsis indicate that components of the SC also facilitate homologous chromosome pairing (Higgins et al. 2005; Tsubouchi and Roeder 2005). Analysis in yeast showed that molecular ZIPper 1 (ZIP1) is required for the coupling of centromeres during early prophase I, and that homologous chromosomes failed to pair in its absence (Tsubouchi and Roeder 2005). Tsubouchi and Roeder (2005) proposed that the role for the coupling was to facilitate homologue pairing by holding chromosomes in close proximity while homology is being assessed. Further support of a role for SC-component proteins in homologous chromosome pairing was provided through analysis of ZYP1 (Arabidopsis homologue of yeast ZIP1) deficient Arabidopsis plants, which displayed recombination between non-homologous chromosomes (Higgins et al. 2005).
Evidence for a functional relationship between recombination and the SC has been provided through the analysis of yeast mutants with decreased activity of SPOrulation 11 (SPO11), a protein essential in the double strand break repair (DSBR) pathway (Henderson and Keeney 2004). In addition to displaying a decreased ability to generate DSBs, spo11 mutants showed corresponding defects in SC formation (Henderson and Keeney 2004). These defects were caused by a decline in the number of ZIP3 complexes, which are thought to represent sites of SC initiation through their role promoting the recruitment of the transverse filament protein ZIP1 (Agarwal and Roeder 2000). Further evidence of a relationship between SC formation and recombination has been provided from C. elegans, S. cerevisiae and Arabidopsis mutants that are defective for a component of the central element (MacQueen et al. 2002; Börner et al. 2004; Higgins et al. 2005). All of these SC-component mutants displayed reduced levels of crossover formation, and immuno-localisation studies indicated that the defects may be caused by a loss of function in correct SC nucleation during leptotene and zygotene. These results therefore indicate a direct link between DSB initiation and SC formation, especially during the early stages of each process.
Recombination
Much of the early and current research on the DSBR pathway was performed in S. cerevisiae which led to the first model of the recombination pathway (Szostak et al. 1983). Since then, this model has been updated as further discoveries have been made (e.g. Allers and Lichten 2001; Bishop and Zickler 2004; Oh et al. 2007). Equivalent, extensive cytological and molecular analysis in plant species (excluding Arabidopsis) has been limited. However, this has not prevented the recombination model to have been used as a framework for Arabidopsis (Sanchez-Moran et al. 2008). Further, through using such models as a starting point, the identification of several orthologues in agricultural crops including wheat, rice, barley, rye (Secale cereale L.) and maize has already been reported (and in some cases these genes have also been characterised) (Franklin et al. 1999; Pawlowski et al. 2003; Jenkins et al. 2005; Li et al. 2007; Khoo et al. 2008; Bovill et al. 2009).
Cytologically, recombination is observed as large multi-protein complexes called recombination nodules (RNs) (Zickler and Kleckner 1998, 1999). These nodules are found closely associated with the SC and are divided into two categories dependent on frequency and size, known as early (ENs) and late recombination nodules (LNs) (reviewed in Anderson and Stack 2005). ENs are associated with axial elements of the SC and appear from leptotene until pachytene, after which they detach from the SC (Stack and Anderson 1986b). Through the use of immuno-localisation data, Anderson and Stack (2005) have assigned recombination proteins with roles in DSB formation and single-end invasion of DNA strands as components of ENs. Based on the functions of the EN components and the large numbers of ENs, the ENs are hypothesised to have roles in searching for DNA homology, chromosome synapsis, and resolving recombination events into crossovers or non-crossovers (Carpenter 1979, 1987, 1988; Stack and Anderson 1986a, 1986b; Zickler and Kleckner 1999; Anderson et al. 2001; Moens et al. 2002). Involvement in the resolution of recombination events into crossovers or non-crossovers is supported by the fact that cytologically observed ENs are replaced by LNs at pachytene. The number of LNs is not only much fewer then ENs (1–6 per bivalent) but also reflective of the crossover frequency per bivalent (Stack et al. 1989; Anderson et al. 2003). Unlike ENs, LNs display interference patterns consistent with crossover interference and their location correlates well with chiasmata (Stack et al. 1989; Anderson et al. 2003). These observations, combined with immuno-localisation of proteins with essential roles in DNA mismatch repair strongly indicate LNs represent crossover sites (Moens et al. 2002).
Mining meiotic mutants in bread wheat: from classical genetics to molecular farming
The classical genetics
Approximately 70% of flowering plants are estimated to have experienced at least one polyploidisation event during their evolution (Bowers et al. 2003). Recent cytological and molecular analysis of other plants, which have previously been considered as diploids, reveals that they may in fact have been allopolyploids which have undergone diploidisation to now exist as paleopolyploids (Shoemaker et al. 1996; Gaut and Doebley 1997; Gomez et al. 1998; Grant et al. 2000; Vision et al. 2000; Gaut 2001). Such a widespread occurrence of this process is reflective of the potential for allopolyploid species to adapt to a wide range of environmental conditions, allowing these plants to survive in adverse environments when compared with their diploid progenitors (reviewed in Ma and Gustafson 2005). Although polyploids contain at least two sets of genomes per cell, such species frequently behave as cytological diploids during meiosis with only homologous chromosomes pairing with one another. The meiotic behaviour of several allopolyploid plant species has been, and continues to be, studied. These include bread wheat (Riley and Chapman 1958; Sears 1982; Martinez et al. 2001; Martinez-Perez et al. 2001; Griffiths et al. 2006; Colas et al. 2008; Boden et al. 2009), oilseed rape (Brassica napus L.) (Attia and Robbelen 1986; Jenczewski et al. 2003; Udall et al. 2005; Leflon et al. 2006; Liu et al. 2006; Nicolas et al. 2008, 2009), oats (Avena sativa L.) (Gauthier and McGinnis 1968; Rajhathy and Thomas 1972), cotton (Gossypium hirsutum L.) (Brown 1954; Reyes-Valdés and Stelly 1995; Ji et al. 2007; Vafaie-Tabar and Chandrashekaran 2007) and tobacco (Nicotiana tabacum L.) (Trojak-Goluch and Berbeć 2003, 2007). Of all these plant species, the hexaploid genome of bread wheat has provided some of the most useful information to date.
Bread wheat is an allohexaploid with three genomes (AABBDD; 2n = 6x = 42) derived from related progenitors. Studies on the origin of bread wheat have indicated that the genome complement formed ~10 000 years ago, through the hybridisation of the D genome progenitor, Triticum tauschii (2n = 2x = 14), and the tetraploid containing the A and B genome, Triticum turgidum (Feldman 2001). While it is well recognised that the progenitor of the A genome is Triticum urartu (2n = 2x = 14), the origin of the B genome is still open to conjecture (reviewed in Feldman and Levy 2005). An important behavioural characteristic of the wheat genome that ensures stabilisation of the hybrid condition is the diploid-like meiotic behaviour, with chromosome pairing occurring exclusively between homologues and not homoeologues. One of the central dangers to the establishment of a new species is the risk of homoeologous chromosome pairing due to the progenitor genomes usually being closely related, as this pairing would reduce fertility and therefore affect the fitness of the new species.
Studies in bread wheat have identified several loci that contribute to the exclusive pairing of bivalents, which ensures the maintenance of homologous chromosome pairing (Riley and Chapman 1958; Wall et al. 1971a; Driscoll 1972; Mello-Sampayo and Canas 1973; Riley and Chapman 1975; Sears 1982). Some of these loci have been termed Pairing homoeologous (Ph), for their ability to suppress interactions between homoeologous chromosomes (Wall et al. 1971b). Two examples are Ph1 and Ph2, which are located on chromosome arms 5BL and 3DS, respectively (Riley and Chapman 1958; Driscoll 1972; Sears 1976).
Of the loci that contribute to the diploid behaviour of meiosis in bread wheat, the Ph1 locus displays the strongest effect (Sears 1976; Feldman 1993; Moore 2002). Since its discovery some 50 years ago, cytological investigations of wheat mutants that lack the Ph1 locus have shown that there are several defects that occur during pre-meiotic interphase and early meiosis that contribute to the homoeologous chromosome pairing observed at metaphase I (Riley and Chapman 1958; Sears 1977; Holm and Wang 1988). Initial studies showed that synapsis is arrested in ph1 mutants, to a level of ~35–40% of wild-type wheat (Holm and Wang 1988). In addition, multiple axial element associations that normally occur during zygotene are not resolved into homologous chromosome pairs in ph1 mutants like they are in wild-type wheat (Holm and Wang 1988). Based on these observations, Holm and Wang (1988) proposed that the alignment of homologous chromosomes is affected in ph1 mutants, preventing the correction of multiple axial element associations. Uncorrected associations are thought to be the prelude to the multivalents observed at metaphase I in ph1 mutants (Holm and Wang 1988). Similarly, Ph1 appears to be required for the synchronous remodelling of homologous chromosomes which occurs at the same time as telomere bouquet formation (Prieto et al. 2004; Colas et al. 2008). The absence of Ph1 causes premature and asynchronous remodelling of homologous chromosomes, which leaves a chromosome just as likely to interact with a related homoeologue as with its true homologue (Martinez-Perez et al. 2001; Prieto et al. 2004; Colas et al. 2008). This has been confirmed by studies showing that Ph1 regulates the specificity of chromosome interactions at sites of centromeres and telomeres, so that pairing only occurs between homologues (Martinez-Perez et al. 2001; Prieto et al. 2004).
In addition to the cytological investigations of meiosis in ph1 mutants, genetic studies have been performed to identify the gene(s) responsible for the effect of the Ph1 locus. The original Ph1 deletion was defined to span a region of 70 Mbp; however, recent studies have reduced the size of this locus to 2.5 Mbp (Gill et al. 1993; Roberts et al. 1999; Griffiths et al. 2006). This refined locus was found to contain seven Cyclin dependent kinase-like (Cdk-like) genes and a segment of sub-telomeric heterochromatin, with the Cdk-like genes shown to be closely related to Cdk2 of human and mouse (Griffiths et al. 2006; Martinez-Perez and Moore 2008). In addition, it has been shown that the Cdk-like genes of the 5B locus suppress the expression of the 5A and 5D Cdk-like loci, which are expressed at higher than normal levels in the absence of Ph1 (Al-Kaff et al. 2008). This suggests that these genes coordinate chromatin remodelling of homologues to ensure that they are in the same conformation at the onset of pairing (Al-Kaff et al. 2008). Taken together, these studies indicate that by investigating the genes and proteins that are affected by deletion of Ph1, it may be possible to understand the mechanism that controls the diploid-like behaviour of bread wheat meiosis. Subsequently, ASY1 localisation has been shown to be perturbed in ph1b mutants (Boden et al. 2009).
Identification of the matchmaker: HOP1/ASY1 is required for chromosome synapsis and homologous chromosome pairing
In 1989, Hollingsworth and Byers identified a yeast gene with roles in SC formation and recombination by screening yeast mutants defective for homologous chromosome pairing during meiosis. Cytological analysis of the hop1 mutant revealed that it failed to form a SC and displayed reduced levels of recombination (Hollingsworth and Byers 1989). Further evidence for a role in such processes was provided by identification of DNA interacting domains within HOP1, as well as immuno-localisation of the protein to sites directly adjacent to the lateral elements of the SC of homologous chromosome pairs (Hollingsworth et al. 1990; Muniyappa et al. 2000; Anuradha and Muniyappa 2004a, 2004b).
Immuno-localisation studies and the identification of proteins that interact with HOP1 also provided evidence for involvement in chromosome synapsis (Hollingsworth et al. 1990; Hollingsworth and Ponte 1997; Woltering et al. 2000). Examination of whole chromosome spreads from meiotic nuclei by electron microscopy following incubation with an anti-HOP1 primary antibody and a colloidal gold-conjugated secondary antibody revealed that the protein is closely associated with meiotic chromosomes (Hollingsworth et al. 1990). While HOP1 was proposed to serve as a component of the SC, comparisons of similar analysis with known SC components from mouse indicates that HOP1 is more likely to represent a non-SC component chromatin interacting factor (Hollingsworth et al. 1990; Schalk et al. 1998). The idea of HOP1 representing a non-SC protein is further supported by its dissociation from chromosomes at pachytene, when SC formation becomes complete (Smith and Roeder 1997).
So what is known about this gene in plant species? As highlighted earlier, identification and isolation of plant meiotic genes via sequence homology with yeast genes can be difficult. Using sequence alone, identifying a HOP1 orthologue in plants proved difficult, and at best returned low percentage matches. To overcome this, plant scientists used a reverse genetics approach by screening Arabidopsis T-DNA insertion lines for plants with reduced fertility and abnormal chromosome behaviour during meiosis. This led to the identification of ASY1, which is required for correct synapsis of homologous chromosomes (Ross et al. 1997; Caryl et al. 2000). Plants with a T-DNA disrupted ASY1 gene display an absence of synapsed bivalents during pachytene, with unconnected homologues appearing as univalents at diplotene and diakinesis (Ross et al. 1997). This is followed by irregular chromosome distribution at metaphase I, and multiple non-disjunction events caused by equational segregation of chromosomes at anaphase I. Such chromosome behaviour led to a reduction in fertility to ~10% of the wild-type (Ross et al. 1997; Caryl et al. 2000). Similar analysis resulted in the identification of the rice ASY1/HOP1 orthologue, termed PAIR2 (Nonomura et al. 2004).
Complementing the forward genetics approach identifying ASY1 and PAIR2 in Arabidopsis and rice, a targeted genetic and protein approach was used to characterise the bread wheat orthologue, TaASY1 (Boden et al. 2007, 2009). TaASY1 displays significant genetic similarities with ASY1 and PAIR2, with expression analysis revealing high levels of transcript at interphase and prophase I of meiosis (Boden et al. 2007). Immuno-gold and immuno-fluorescence localisation of TaASY1 in meiotic cells further supports a conserved role for this protein, with analysis in all three organisms showing that ASY1/PAIR2 localises to chromatin of lateral elements of the SC as chromosomes are condensing and pairing with their homologous partner (Armstrong et al. 2001; Nonomura et al. 2006; Boden et al. 2007, 2009). These analyses also reported a strict temporal localisation pattern for ASY1/PAIR2; with the protein first localising to chromatin at early leptotene as punctate foci that polymerise to form a continuous signal throughout zygotene, before dissociating from the chromatin at the completion of synapsis, so that the signal diminishes during pachytene to be absent by diakinesis (Fig. 3). This suggested that ASY1/PAIR2 was required for either synapsis, by recruiting proteins to form the structures required for formation of the SC, or pairing by facilitating the chromosomes’ search for their homologous partner.
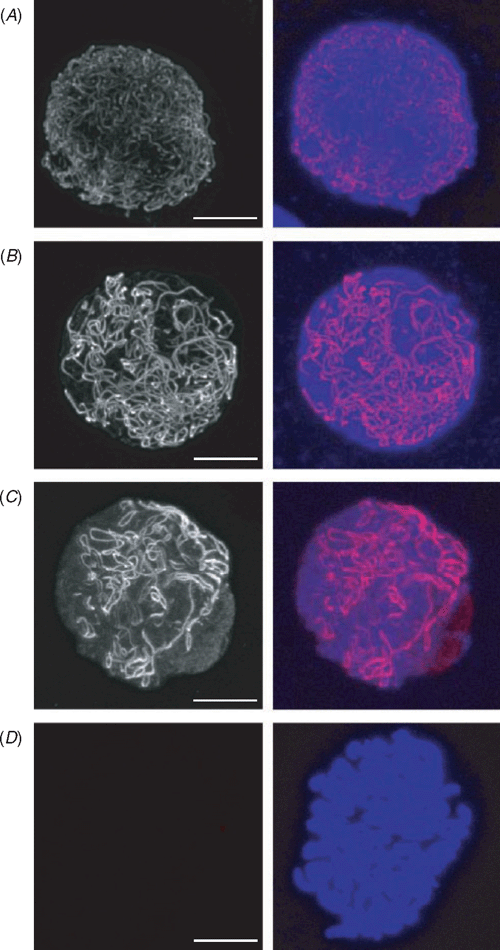
|
Recent analyses performed in Taasy1 RNA interference (RNAi) bread wheat lines and Chinese Spring ph1b bread wheat have indicated that a role in chromosome pairing is more likely. In the Taasy1 lines with reduced levels of TaASY1 transcript, the strict control of chromosome pairing between homologues was reduced, such that multivalents and univalents were observed at metaphase I (Boden et al. 2009). The metaphase I phenotype was reminiscent of that from ph1 deficient lines previously reported (Sears 1977; Fig. 4). Transcript analysis of TaASY1 in ph1b showed a significant 20-fold increase relative to wild-type. Interestingly, this observation is consistent with recent observations of HOP1 gene activity in yeast meiosis following deletion of the Cdk2 homologue, named Inducer of meiosis 2 (Ime2) (Szwarcwort-Cohen et al. 2009), indicating the mechanism controlling chromosome pairing is conserved across eukaryotes. Additional evidence for TaASY1 being involved in bread wheat chromosome pairing was shown by Boden et al. (2009) with the localisation patterns in the ph1b mutant being disrupted and spiral-like in appearance. This observation is consistent with abnormalities in chromatin re-modelling and synapsis that are thought to lead to the homoeologous interactions observed in this mutant (Holm and Wang 1988; Prieto et al. 2004). Combined, these recent findings indicate that TaASY1 is intimately involved with the Ph1 dependent control of chromosome pairing, and that it might promote pairing between homologous regions of chromosomes. This activity is being controlled at a transcriptional level by Ph1 in wild-type wheat so that regions of close homology (e.g. homoeologous sequences) are not promoted to interact with each other. An updated model illustrating how these new results contribute to understanding homologue pairing interactions during wheat meiosis was recently published (Moore and Shaw 2009).
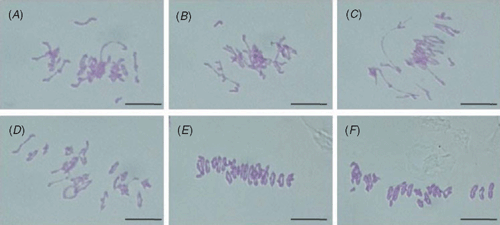
|
Molecular farming to discover other orthologues and novel candidates
In the plant kingdom, Arabidopsis remains the favoured model organism because of its short life cycle, small size and small genome. With the genome having been sequenced in 2000, and the availability of an extensive T-DNA mutant collection with flanking sequence tags (Samson et al. 2002; Sessions et al. 2002; Alonso et al. 2003), extensive research on a gene of choice can be completed in a relatively short period of time when compared with some of the larger genomes such as barley and bread wheat. In a little over a decade of Arabidopsis research, various research groups have identified and characterised ~50 genes with roles in meiosis (Mercier and Grelon 2008). This list, while not exhaustive when compared with what has been reported in yeast, is comprised of a mixture of novel genes and those with orthologues in other taxonomic kingdoms.
While the sequencing and assembly of the barley and bread wheat genomes are in their formative years, comparative genetics using the rice genome sequence has enabled several regions that share synteny in these genomes to be identified (Sutton et al. 2003; Griffiths et al. 2006; Paux et al. 2006; Jardim 2007; Huang et al. 2008; March et al. 2008). One of the most appropriate studies to discuss here is the research conducted with the Ph1 locus. The similarity between bread wheat (genome size of ~17 000 Mbp), rice (~490 Mbp) and Brachypodium sylvaticum (Huds.) Beauv. (~160 Mbp) genomes were used to create the framework for the bacterial artificial chromosomes (BACs) that contained sequence from the Ph1 locus and also the homoeologous regions on the long arms of chromosomes 5A and 5D (Griffiths et al. 2006). This enabled the original fast-neutron irradiation-induced 70 Mbp deletion to be refined to a 2.5 Mbp region, and the identification of genes believed to be responsible for the action of Ph1. Using a similar approach Sutton et al. (2003) scrutinised the gene content of the Ph2 locus, which as highlighted earlier, is another region that influences chromosome pairing behaviour in bread wheat. In identifying 280 expressed sequence tags (ESTs) from rice chromosome 1 which was 6.58 Mbp in length and syntenic to the Ph2 locus, Sutton et al. (2003) mapped a subset of these ESTs and showed that 78% were located within the Ph2 region. Information pertaining to putative genes within this region that may encode the action of Ph2 was also reported.
More recently, a comparative genetics approach was used to identify meiotic gene candidates in the small grain cereals using the yeast and Arabidopsis sequenced genomes (Bovill et al. 2009). From 53 genes known to be involved in meiosis in either yeast and/or Arabidopsis, 30 or more orthologues from wheat, rice and barley were identified with an E-value > E–20. This collection of ESTs (and full-length sequences in some instances) clearly demonstrates the level of DNA conservation across diverse organisms and is a valuable resource from which further studies can be launched, including transgenic studies of individual candidates in the cereals (see Table 2 for examples). As the diversity of technologies used for meiotic research increases, there will also be comparisons made between biological processes such as the transcriptomes of organisms and the biochemical pathways that they control. Indeed, one approach used to extract meaningful outcomes from vast amounts of data involves the constantly evolving microarray technology; initially designed as a high throughput platform to quantify gene expression (Schena et al. 1995).
Since its inception, reproductive processes including stages of meiosis, differences in germline and somatic tissues, and differences in male and female germlines have been investigated using microarrays. However not surprisingly, the majority of these studies were completed in model species including; S. cerevisiae, Schizosaccharomyces pombe (fission yeast), C. elegans, D. melanogaster, rats (Rattus rattus), and Arabidopsis (Chu et al. 1998; Andrews et al. 2000; Primig et al. 2000; Reinke et al. 2000; Mata et al. 2002; Schlecht et al. 2004). Some of the earliest meiotic microarray work investigated the transcriptional program of S. cerevisiae (Chu et al. 1998). Complementary DNA microarrays containing 97% of the known S. cerevisiae genes were used to increase the number of genes meiotically-regulated from ~150 which were identified using conventional methods (Chu et al. 1998; and references therein), to over 1000 using a microarray approach. Subsequently the two S. cerevisiae strains, SK1 and W303, which show different sporulation efficiencies, were compared. This revealed gene deletions, polymorphisms and ~1600 temporally-regulated genes in both strains (Primig et al. 2000). These genes were assigned into seven broad expression clusters; with some of them having previously been reported in DNA synthesis, recombination, the synaptonemal complex and sporulation. Of significance in this study was the identification of ~650 meiotically-regulated genes not previously mentioned in the literature (Primig et al. 2000).
More recently, Crismani et al. (2006) analysed a large-scale transcriptomics dataset across a meiotic time series in bread wheat. They showed that 1350 transcripts were temporally-regulated during the early stages of meiosis, in which 30 of these had at least an 8-fold expression change between different stages. While a significant proportion of the 1350 transcripts hit to uncharacterised sequences or did not share any sequence similarity with any database entry to date, several shared sequence similarity with genes which have roles in chromatin condensation, synaptonemal complex formation and recombination (Crismani et al. 2006). Where limited sequence information for the organism of choice exists, microarray analysis has therefore proven useful in identifying novel (and known) meiosis candidates to undertake further research on. Evidence that wide-scale synergies exist between the expression profiles of meiosis genes from wheat, poplar (Populus trichocarpa Torr. & Gray) and Arabidopsis have also been discovered (W. Crismani and J. A. Able, unpubl. data). Using comparative expression profiling may therefore be an additional approach to identify important meiotic genes and/or gene clusters between similar and/or divergent species of interest.
Implications for crop improvement: why manipulate meiosis?
For the majority of crop species we have access to genetic variation in the cultivated gene pool, amongst land races and in the wild relatives. Although these sources have been used for crop improvement, plant breeders have typically relied upon exploiting genetic variation present within cultivated and well adapted lines in order to improve their target phenotypes and to minimise the transfer of unwanted linkage blocks. As highlighted earlier in the case of bread wheat, recombination of wild chromatin into the genetic makeup of these elite lines is strongly suppressed and is a direct result of the tight meiotic controls prevalent in this species. As breeders continually face commercial pressures to generate new varieties quickly, this has meant that they are often reluctant to use poorly adapted lines, land races and wild relatives due to the slow and complex introgression of the desired alleles into commercial varieties. Given that speciation of many crops has occurred over thousands of years, the level of natural variability is invariably large. Just how much of this diversity has been captured, especially within the small cereal grain families, remains unclear. Estimates suggest that in the Triticeae alone, only ~10 to 15% of the gene pool has been utilised (Able and Langridge 2006). Significantly expanding the use of non-adapted and wild germplasm in breeding programs will therefore depend on the ability to manipulate key events such as meiotic recombination. With a greater ability to regulate the location and/or frequency of recombination events, undesirable linkage drag would be minimised in any particular cross. Reducing linkage drag would subsequently enhance the rate of genetic gain that breeding programs achieve, as target genes could be transferred independently of undesirable genes. Furthermore, producing highly recombinogenic lines would enable plant breeders to reduce the size of populations used, while still obtaining the desired phenotypes.
While major advancements have been made over the past 50 years in various model and agriculturally important crop species, many key events during meiosis are far from being completely understood. This is because meiotic biochemical pathways operate through an integrated and complicated array of networks that are yet to be deciphered. This review has highlighted the importance of meiosis and the benefits of manipulating the key events of chromosome pairing and recombination, with a particular emphasis on bread wheat. From an applied perspective, and as described by Riley and Chapman (1958) with the discovery of Ph1, identification of the mechanisms that control homologous chromosome pairing in bread wheat would have both theoretical and practical implications. Future studies in bread wheat may indeed help to identify proteins that are involved in homology searching during prophase I in a way that is not possible in diploid organisms. In addition, by understanding the mechanisms that suppress homoeologous chromosome interactions, it should be possible to manipulate this process so that homoeologous chromosomes can interact in inter-specific hybrids, which would in turn facilitate the introduction of alien genes into wheat chromosomes by normal recombination. Given the recent research reported by Boden et al. (2009), the relationship between key genes such as TaASY1 and the Ph1 locus is now becoming more complete. Further research investigating other candidates in the Ph1 background is underway and will no doubt lead to a more comprehensive dissection of how chromosomes pair and recombination events take place in this complex polyploid. Such knowledge will have significant implications for plant breeding strategies of the future.
Indeed, since the early 1990s, the adoption of technology such as marker assisted selection and other related marker-based strategies have enabled significant improvements in plant breeding programs to occur. Through further refinements of these current platforms and the development of further innovative assays, the identification and function(s) of new genes and proteins that the pre- and plant breeder will have at their disposal is virtually immeasurable. While some time exists before outcomes of the current ‘omics’ find an applied marker route, such reverse genetics strategies should also eventually find a path through to breeding programs. The power of all these modern day approaches, combined with classical knowledge will ultimately underpin the ability to successfully manipulate the meiotic process not only in commodities such as bread wheat but many crop species of agricultural importance.
Acknowledgements
This research was supported in part by the Molecular Plant Breeding Cooperative Research Centre (MPB CRC), the Grains Research and Development Corporation (GRDC), and the Australian Government under the Australia–India Strategic Research Fund (AISRF). The authors thank Kelvin Khoo for the images that contributed to Fig. 1. We apologise to those colleagues who have contributed to this field but were not cited due to space constraints and thank the two anonymous referees who provided useful feedback and suggestions for improving the content of the review.
Able JA, Langridge P
(2006) Wild sex in the grasses. Trends in Plant Science 11, 261–263.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Able JA,
Langridge P, Milligan AS
(2007) Capturing diversity in the cereals: many options but little promiscuity. Trends in Plant Science 12, 71–79.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Agarwal S, Roeder GS
(2000) Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102, 245–255.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Agashe B,
Prasad CK, Siddiqi I
(2002) Identification and analysis of DYAD: a gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129, 3935–3943.
|
CAS |
PubMed |

Al-Kaff N,
Knight E,
Bertin I,
Foote T,
Hart N,
Griffiths S, Moore G
(2008) Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: with deletion mutants and expression profiling. Annals of Botany 101, 863–872.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Albini SM, Jones GH
(1988) Synaptonemal complex spreading in Allium cepa and Allium fistulosum. II Pachytene observations. The SC karyotype and the correspondence of late recombination nodules and chiasmata. Genome 30, 399–410.

Allers T, Lichten M
(2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Alonso JM,
Stepanova AN,
Leisse TJ,
Kim CJ, Chen HM ,
et al
.
(2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Anderson LK, Stack SM
(2005) Recombination nodules in plants. Cytogenetic and Genome Research 109, 198–204.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Anderson LK,
Hooker KD, Stack SM
(2001) The distribution of early recombination nodules on zygotene bivalents from plants. Genetics 159, 1259–1269.
|
CAS |
PubMed |

Anderson LK,
Doyle GG,
Brigham B,
Carter J,
Hooker KD,
Lai A,
Rice M, Stack SM
(2003) High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165, 849–865.
|
CAS |
PubMed |

Andrews J,
Bouffard GG,
Cheadle C,
Lu JN,
Becker KG, Oliver B
(2000) Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Research 10, 2030–2043.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Anuradha S, Muniyappa K
(2004a) Meiosis-specific yeast Hop1 protein promotes synapsis of double-stranded DNA helices via the formation of guanine quartets. Nucleic Acids Research 32, 2378–2385.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Anuradha S, Muniyappa K
(2004b)
Saccharomyces cerevisiae Hop1 zinc finger motif is the minimal region required for its function in vitro. The Journal of Biological Chemistry 279, 28961–28969.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Armstrong SJ,
Franklin FCH, Jones GH
(2001) Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. Journal of Cell Science 114, 4207–4217.
|
CAS |
PubMed |

Armstrong SJ,
Caryl AP,
Jones GH, Franklin FCH
(2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. Journal of Cell Science 115, 3645–3655.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Attia T, Robbelen G
(1986) Meiotic pairing in haploids and amphidiploids of spontaneous versus synthetic origin in rape, Brassica napus L. Canadian Journal of Genetics and Cytology 28, 330–334.

Bishop DK, Zickler D
(2004) Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117, 9–15.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Boden SA,
Shadiac N,
Tucker EJ,
Langridge P, Able JA
(2007) Expression and functional analysis of TaASY1 during meiosis of bread wheat (Triticum aestivum). BMC Molecular Biology 8, 65.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Boden SA,
Langridge P,
Spangenberg G, Able JA
(2009) TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. The Plant Journal 57, 487–497.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Börner GV,
Kleckner N, Hunter N
(2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Bovill WD,
Deveshwar P,
Kapoor S, Able JA
(2009) Whole genome approaches to identify early meiotic gene candidates in cereals. Functional & Integrative Genomics 9, 219–229.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Bowers JE,
Chapman BA,
Rong J, Paterson AH
(2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Brown MS
(1954) A comparison of pachytene and metaphase pairing in species hybrids of Gossypium. Genetics 39, 962–963.

Carpenter A
(1975) Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement and temporal change of the synaptonemal complex in wild-type. Chromosoma 51, 157–182.
|
CAS |
Crossref |
PubMed |

Carpenter T
(1979) Synaptonemal complex and recombination nodules in wild type Drosophila melanogaster females. Genetics 92, 511–541.
|
CAS |
PubMed |

Carpenter T
(1987) Gene conversion, recombination nodules, and the initiation of meiotic synapsis. BioEssays 6, 232–236.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Caryl AP,
Armstrong SJ,
Jones GH, Franklin FCH
(2000) A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109, 62–71.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Chu S,
DeRisi J,
Eisen M,
Mulholland J,
Botstein D,
Brown PO, Herskowitz I
(1998) The transcriptional program of sporulation in budding yeast. Science 282, 699–705.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Colas I,
Shaw P,
Prieto P,
Wanous M,
Spielmeyer W,
Mago R, Moore G
(2008) Effective chromosome pairing requires chromatin remodeling at the onset of meiosis. Proceedings of the National Academy of Sciences of the United States of America 105, 6075–6080.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Crismani W,
Baumann U,
Sutton T,
Shirley N,
Webster T,
Spangenberg G,
Langridge P, Able JA
(2006) Microarray expression analysis of meiosis and microsporogenesis in hexaploid bread wheat. BMC Genomics 7, 267.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Driscoll C
(1972) Genetic suppression of homologous chromosome pairing in hexaploid wheat. Canadian Journal of Genetics and Cytology 14, 39–42.

Feldman M
(1993) Cytogenetic activity and mode of action of the pairing homoeologous (Ph1) gene of wheat. Crop Science 33, 894–897.

Feldman M, Levy AA
(2005) Allopolyploidy – a shaping force in the evolution of wheat genomes. Cytogenetic and Genome Research 109, 250–258.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Franklin AE,
McElver J,
Sunjevaric I,
Rothstein R,
Bowen B, Cande WZ
(1999) Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. The Plant Cell 11, 809–824.
|
CAS |
Crossref |
PubMed |

Fung J,
Marshall W,
Dernburg A,
Agard D, Sedat J
(1998) Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. The Journal of Cell Biology 141, 5–20.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Gaut BS
(2001) Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Research 11, 55–66.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Gaut BS, Doebley JF
(1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proceedings of the National Academy of Sciences of the United States of America 94, 6809–6814.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Gauthier FM, McGinnis RC
(1968) The meiotic behaviour of a nullihaploid plant in Avena sativa L. Canadian Journal of Genetics and Cytology 10, 186–189.

Gill KS,
Gill BS,
Endo TR, Mukai Y
(1993) Fine physical mapping of Ph1, a chromosome-pairing regulator gene in polyploid wheat. Genetics 134, 1231–1236.
|
CAS |
PubMed |

Goldstein P, Slaton D
(1982) Synaptonemal complexes of Caenorhabditis elegans: comparison of wild-type and mutant strains and pachytene karyotype analysis of wild-type. Chromosoma 84, 585–597.
|
CAS |
Crossref |
PubMed |

Golubovskaya IN,
Harper LC,
Pawlowski WP,
Schichnes D, Cande WZ
(2002) The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162, 1979–1993.
|
CAS |
PubMed |

Gomez MI,
Islam-Faridi MN,
Zwick MS,
Czeschin DG,
Hart GE,
Wing RA,
Stelly DM, Price HJ
(1998) Tetraploid nature of Sorghum bicolor (L.) Moench. The Journal of Heredity 89, 188–190.
| Crossref | GoogleScholarGoogle Scholar |

Grant D,
Cregan P, Shoemaker RC
(2000) Genome organisation in dicots: Genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 97, 4168–4173.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Griffiths S,
Sharp R,
Foote TN,
Bertin I,
Wanous M,
Reader S,
Colas I, Moore G
(2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439, 749–752.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hamant O,
Ma H, Cande WZ
(2006) Genetics of meiotic prophase I in plants. Annual Review of Plant Biology 57, 267–302.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Harper L,
Golubovskaya I, Cande WZ
(2004) A bouquet of chromosomes. Journal of Cell Science 117, 4025–4032.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Henderson KA, Keeney S
(2004) Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proceedings of the National Academy of Sciences of the United States of America 101, 4519–4524.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Higgins JD,
Sanchez-Moran E,
Armstrong SJ,
Jones GH, Franklin FCH
(2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes & Development 19, 2488–2500.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hiraoka T
(1952) Observational and experimental studies of meiosis with special reference to the bouquet stage. VIII. An experimental study on the cell and nuclear conditions in the bouquet stage. Cytologia 16, 335–340.

Hollingsworth NM, Byers B
(1989) Hop1 – A yeast meiotic pairing gene. Genetics 121, 445–462.
|
CAS |
PubMed |

Hollingsworth NM, Ponte L
(1997) Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics 147, 33–42.
|
CAS |
PubMed |

Hollingsworth NM,
Goetsch L, Byers B
(1990) The Hop1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61, 73–84.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Holm P, Wang X
(1988) The effect of chromosome 5B on synapsis and chiasma formation in wheat Triticum aestivum cv. Chinese Spring. Carlsberg Research Communications 53, 191–208.
| Crossref | GoogleScholarGoogle Scholar |

Huang S,
Spielmeyer W,
Lagudah ES, Munns R
(2008) Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. Journal of Experimental Botany 59, 927–937.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hunter N
(2003) Synaptonemal complexities and commonalities. Molecular Cell 12, 533–535.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Jardim SN
(2007) Comparative genomics of grasses tolerant to aluminum. Genetics and Molecular Research 6, 1178–1189.
|
CAS |
PubMed |

Jenczewski E,
Eber F,
Grimaud A,
Huet S,
Lucas MO,
Monod H, Chevre AM
(2003)
PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164, 645–653.
|
CAS |
PubMed |

Jenkins G,
Mikhailova EI,
Langdon T,
Tikholiz OA,
Sosnikhina SP, Jones RN
(2005) Strategies for the study of meiosis in rye. Cytogenetic and Genome Research 109, 221–227.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ji YF,
Zhao XP,
Paterson AH,
Price HJ, Stelly DM
(2007) Integrative mapping of Gossypium hirsutum L. by meiotic fluorescent in situ hybridization of a tandemly repetitive sequence (B77). Genetics 176, 115–123.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Khoo KHP,
Jolly HR, Able JA
(2008) The RAD51 gene family in bread wheat is highly conserved across eukaryotes, with RAD51A upregulated during early meiosis. Functional Plant Biology 35, 1267–1277.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Kleckner N
(1996) Meiosis: how could it work? Proceedings of the National Academy of Sciences of the United States of America 93, 8167–8174.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Kleckner N
(2006) Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Klein F,
Mahr P,
Galova M,
Buonomo SBC,
Michaelis C,
Nairz K, Nasmyth K
(1999) A central role for cohesions in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Leflon M,
Eber F,
Letanneur JC,
Chelysheva L,
Coriton O,
Huteau V,
Ryder CD,
Barker G,
Jenczewski E, Chevre AM
(2006) Pairing and recombination at meiosis of Brassica rapa (AA) × Brassica napus (AACC) hybrids. Theoretical and Applied Genetics 113, 1467–1480.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Li J,
Harper LC,
Golubovskaya IN,
Wang CR,
Weber D,
Meeley RB,
McElver J,
Bowen B,
Cande WZ, Schnable PS
(2007) Functional analysis of maize RAD51 in meiosis and double-strand break repair. Genetics 176, 1469–1482.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Liu ZQ,
Adamczyk K,
Manzanares-Dauleux M,
Eber F,
Lucas MO,
Delourme R,
Chevre AM, Jenczewski E
(2006) Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 174, 1583–1596.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Lloyd RG, Sharples GJ
(1993) Dissociation of synthetic Holliday junctions by E. coli RecG protein. The EMBO Journal 12, 17–22.
|
CAS |
PubMed |

Lloyd AH,
Milligan AS,
Langridge P, Able JA
(2007)
TaMSH7: a cereal mismatch repair gene that affects fertility in transgenic barley (Hordeum vulgare L.). BMC Plant Biology 7, 67.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ma XF, Gustafson JP
(2005) Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenetic and Genome Research 109, 236–249.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

MacQueen AJ,
Colaiacovo MP,
McDonald K, Villeneuve AM
(2002) Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & Development 16, 2428–2442.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

MacQueen AJ,
Phillips CM,
Bhalla N,
Weiser P,
Villeneuve AM, Dernburg AF
(2005) Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123, 1037–1050.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

March TJ,
Able JA,
Willsmore K,
Schultz CJ, Able AJ
(2008) Comparative mapping of a QTL controlling black point formation in barley. Functional Plant Biology 35, 427–437.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Martinez M,
Cunado N,
Carcelen N, Romero C
(2001) The Ph1 and Ph2 loci play different roles in the synaptic behaviour of hexaploid wheat Triticum aestivum. Theoretical and Applied Genetics 103, 398–405.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Martinez-Perez E, Moore G
(2008) To check or not to check? The application of meiotic studies to plant breeding. Current Opinion in Plant Biology 11, 222–227.
| PubMed |

Martinez-Perez E,
Shaw P, Moore G
(2001) The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature 411, 204–207.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Mata J,
Lyne R,
Burns G, Bahler J
(2002) The transcriptional program of meiosis and sporulation in fission yeast. Nature Genetics 32, 143–147.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Mercier R, Grelon M
(2008) Meiosis in plants: ten years of gene discovery. Cytogenetic and Genome Research 120, 281–290.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Moens PB,
Kolas NK,
Tarsounas M,
Marcon E,
Cohen PE, Spyropoulos B
(2002) The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. Journal of Cell Science 115, 1611–1622.
|
CAS |
PubMed |

Moore G
(2002) Meiosis in allopolyploids – the importance of ‘Teflon’ chromosomes. Trends in Genetics 18, 456–463.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Moore G, Shaw P
(2009) Improving the chances of finding the right partner. Current Opinion in Genetics & Development 19, 99–104.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Moses M
(1968) Synaptonemal complex. Annual Review of Genetics 2, 363–412.
| Crossref | GoogleScholarGoogle Scholar |

Motamayor JC,
Vezon D,
Bajon C,
Sauvanet A,
Grandjean O,
Marchand M,
Bechtold N,
Pelletier G, Horlow C
(2000)
Switch (swi1), an Arabidopsis thaliana mutant affected in the female meiotic switch. Sexual Plant Reproduction 12, 209–218.
| Crossref | GoogleScholarGoogle Scholar |

Muniyappa K,
Anuradha S, Byers B
(2000) Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Molecular and Cellular Biology 20, 1361–1369.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Nicolas SD,
Leflon M,
Liu Z,
Eber F,
Chelysheva L,
Coriton O,
Chèvre AM, Jenczewski E
(2008) Chromosome ‘speed dating’ during meiosis of polyploid Brassica hybrids and haploids. Cytogenetic and Genome Research 120, 331–338.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Nicolas SD,
Leflon M,
Monod H,
Eber F,
Coriton O,
Huteau V,
Chèvre AM, Jenczewski E
(2009) Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. The Plant Cell ,
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Nonomura K,
Nakano M,
Murata K,
Miyoshi K,
Eiguchi M,
Miyao A,
Hirochika H, Kurata N
(2004) An insertional mutation in the rice PAIR2 gene, the orthologue of ArabidopsisASY1, results in a defect in homologous chromosome pairing during meiosis. Molecular Genetics and Genomics 271, 121–129.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Nonomura K,
Nakano M,
Eiguchi M,
Suzuki T, Kurata N
(2006) PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. Journal of Cell Science 119, 217–225.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Novak JE,
Ross-Macdonald PB, Roeder GS
(2001) The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158, 1013–1025.
|
CAS |
PubMed |

Oh SD,
Lao JP,
Hwang PY,
Taylor AF,
Smith GR, Hunter N
(2007) BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Orr-Weaver T
(1995) Meiosis in Drosophila: seeing is believing. Proceedings of the National Academy of Sciences of the United States of America 92, 10443–10449.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Page S, Hawley R
(2004) The genetics and molecular biology of the synaptonemal complex. Annual Review of Cell and Developmental Biology 20, 525–558.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Paux E,
Roger D,
Badaeva E,
Gay G,
Bernard M,
Sourdille P, Feuillet C
(2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. The Plant Journal 48, 463–474.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Pawlowski WP,
Golubovskaya IN, Cande WZ
(2003) Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. The Plant Cell 15, 1807–1816.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Pawlowski WP,
Golubovskaya IN,
Timofejeva L,
Meeley RB,
Sheridan WF, Cande WZ
(2004) Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303, 89–92.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Prieto P,
Shaw P, Moore G
(2004) Homologue recognition during meiosis is associated with a change in chromatin conformation. Nature Cell Biology 6, 906–908.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Primig M,
Williams RM,
Winzeler EA,
Tevzadze GG,
Conway AR,
Hwang SY,
Davis RW, Esposito RE
(2000) The core meiotic transcriptome in budding yeasts. Nature Genetics 26, 415–423.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Rajhathy T, Thomas H
(1972) Genetic control of chromosome pairing in hexaploid oats. Nature: New Biology 239, 217–219.
|
CAS |
Crossref |
PubMed |

Reinke V,
Smith HE,
Nance J,
Wang J, Van Doren C ,
et al
.
(2000) A global profile of germline gene expression in C. elegans. Molecular Cell 6, 605–616.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Reyes-Valdés MH, Stelly DM
(1995) A maximum-likelihood algorithm for genome mapping of cytogenetic loci from meiotic configuration data. Proceedings of the National Academy of Sciences of the United States of America 92, 9824–9828.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Riley R, Chapman V
(1958) Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182, 713–715.
| Crossref | GoogleScholarGoogle Scholar |

Riley R, Chapman V
(1975) Genetic control of the cytological diploid behaviour of hexaploid wheat. Nature Reviews. Molecular Cell Biology 182, 712–715.

Roberts M,
Reader S,
Dalgliesh C,
Miller T,
Foote T,
Fish L,
Snape J, Moore G
(1999) Induction and characterisation of Ph1 wheat mutants. Genetics 153, 1909–1918.
|
CAS |
PubMed |

Rockmill B,
Sym M,
Scherthan H, Roeder GS
(1995) Roles for 2 RecA homologs in promoting meiotic chromosome synapsis. Genes & Development 9, 2684–2695.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ross K,
Fransz P,
Armstrong S,
Vizir I,
Mulligan B,
Franklin F, Jones G
(1997) Cytological characterisation of four meiotic mutants of Arabidopsis isolated from T-DNA transformed lines. Chromosome Research 5, 551–559.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Samson SF,
Brunaud V,
Balzergue S,
Dubreucq B,
Lepiniec L,
Pelletier G,
Caboche M, Lecharny A
(2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Research 30, 94–97.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Sanchez-Moran E,
Santos JL,
Jones GH, Franklin FCH
(2007) ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes & Development 21, 2220–2233.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Sanchez-Moran E,
Osman K,
Higgins JD,
Pradillo M,
Cunado N,
Jones GH, Franklin FCH
(2008) ASY1 coordinates early events in the plant meiotic recombination pathway. Cytogenetic and Genome Research 120, 302–312.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Schalk JAC,
Dietrich AJJ,
Vink ACG,
Offenberg HH,
van Aalderen M, Heyting C
(1998) Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma 107, 540–548.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Schena M,
Shalon D,
Davis RW, Brown PO
(1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Schlecht U,
Demougin P,
Koch R,
Hermida L,
Wiederkehr C,
Descombes P,
Pineau C,
Jegou B, Primig M
(2004) Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Molecular Biology of the Cell 15, 1031–1043.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Sears E
(1976) Genetic control of chromosome pairing in wheat. Annual Review of Genetics 10, 31–51.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Sears E
(1977) An induced mutant with homoeologous pairing in common wheat. Canadian Journal of Genetics and Cytology 19, 585–593.

Sears E
(1982) A wheat mutation conditioning an intermediate level of homoeologous chromosome pairing. Canadian Journal of Genetics and Cytology 24, 715–719.

Sessions A,
Burke E,
Presting G,
Aux G, McElver J ,
et al
.
(2002) A high-throughput Arabidopsis reverse genetics system. The Plant Cell 14, 2985–2994.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Shoemaker RC,
Polzin K,
Labate J,
Specht J, Brummer EC ,
et al
.
(1996) Genome duplication in soybean (Glycine subgenus soja). Genetics 144, 329–338.
|
CAS |
PubMed |

Smith A, Roeder G
(1997) The yeast Red1 protein localizes to the cores of meiotic chromosomes. The Journal of Cell Biology 136, 957–967.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Stack SM, Anderson LK
(1986a) The relation between synapsis, the synaptonemal complex, and crossing over in Zea mays. American Journal of Botany 73, 687–688.
| Crossref |

Stack SM, Anderson LK
(1986b) Two-dimensional spreads of synaptonemal complexes from solanaceous plants. II Synapsis in Lycopersicon esculentum (Tomato). American Journal of Botany 73, 264–281.
| Crossref | GoogleScholarGoogle Scholar |

Stack SM,
Anderson LK, Sherman JD
(1989) Chiasmata and recombination nodules in Lilium longiflorum. Genome 32, 486–498.

Sutton T,
Whitford R,
Baumann U,
Dong CM,
Able JA, Langridge P
(2003) The Ph2 pairing homoeologous locus of wheat (Triticum aestivum): identification of candidate meiotic genes using a comparative genetics approach. The Plant Journal 36, 443–456.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Szostak JW,
Orrweaver TL,
Rothstein RJ, Stahl FW
(1983) The double-strand break repair model for recombination. Cell 33, 25–35.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Szwarcwort-Cohen M,
Kasulin-Boneh Z,
Sagee S, Kassir Y
(2009) Human Cdk2 is a functional homolog of budding yeast Ime2, the meiosis-specific Cdk-like kinase. Cell Cycle (Georgetown, Tex.) 8, 647–654.
|
CAS |
PubMed |

Trojak-Goluch A, Berbeć A
(2003) Cytological investigations of the interspecific hybrids of Nicotiana tabacum L. × N. glauca Grah. Journal of Applied Genetics 44, 45–54.
| PubMed |

Trojak-Goluch A, Berbeć A
(2007) Meiosis and fertility in interspecific hybrids of Nicotiana tabacum L. × N. glauca Grah. and their derivatives. Plant Breeding 126, 201–206.
| Crossref | GoogleScholarGoogle Scholar |

Tsubouchi T, Roeder GS
(2005) A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308, 870–873.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Udall JA,
Quijada PA, Osborn TC
(2005) Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics 169, 967–979.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Vafaie-Tabar M, Chandrashekaran S
(2007) Meiosis in a triploid hybrid of Gossypium: high frequency of secondary bipolar spindles at metaphase II. Journal of Genetics 86, 45–49.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Vision TJ,
Brown DG, Tanksley SD
(2000) The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

von Wettstein D,
Rasmussen S, Holm P
(1984) The synaptonemal complex in genetic segregation. Annual Review of Genetics 18, 331–413.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Wall A,
Riley R, Chapman V
(1971a) Wheat mutants permitting homoeologous meiotic chromosome pairing. Genetical Research 18, 311–328.

Wall A,
Riley R, Gale M
(1971b) The position of a locus on chromosome 5B of Triticum aestivum affecting homoeologous meiotic pairing. Genetical Research 18, 329–339.

Whitby MC, Lloyd RG
(1995) Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. The EMBO Journal 14, 3302–3310.
|
CAS |
PubMed |

Woltering D,
Baumgartner B,
Bagchi S,
Larkin B,
Loidl J,
de los Santos T, Hollingsworth N
(2000) Meiotic segregation, synapsis and recombination checkpoint functions require physical interaction between chromosomal proteins Red1p and Hop1p. Molecular and Cellular Biology 20, 6646–6658.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Yu HG, Koshland DE
(2003) Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. The Journal of Cell Biology 163, 937–947.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Zickler D, Kleckner N
(1998) The leptotene-zygotene transition of meiosis. Annual Review of Genetics 32, 619–697.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Zickler D, Kleckner N
(1999) Meiotic chromosomes: integrating structure and function. Annual Review of Genetics 33, 603–754.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |




