Identification and functional characterisation of aquaporins in the grapevine, Vitis vinifera
Megan C. Shelden A B , Susan M. Howitt C , Brent N. Kaiser A and Stephen D. Tyerman A DA School of Agriculture, Food and Wine, University of Adelaide, Waite Campus, Glen Osmond, SA 5064, Australia.
B Present address: Australian Centre for Plant Functional Genomics, School of Botany, University of Melbourne, Parkville, Vic. 3010, Australia.
C Biochemistry and Molecular Biology, Research School of Biology, Australian National University, Canberra, ACT 0200, Australia.
D Corresponding author. Email: stephen.tyerman@adelaide.edu.au
Functional Plant Biology 36(12) 1065-1078 https://doi.org/10.1071/FP09117
Submitted: 21 May 2009 Accepted: 28 July 2009 Published: 3 December 2009
Abstract
Plant aquaporins belong to a large superfamily of conserved proteins called the major intrinsic proteins (MIPs). There is limited information about the diversity of MIPs in grapevine, and their water transport capacity. The aim of the present study was to identify MIPs from grapevine and functionally characterise water transport of a subset of MIPs. Candidate genes were identified, by screening a Vitis vinifera L. (cv. Cabernet Sauvignon) cDNA library with gene specific probes, for aquaporin cDNAs encoding members of the plasma membrane intrinsic protein (PIP) and tonoplast intrinsic protein (TIP) subfamilies. The screen resulted in the identification of 11 full-length and two partial length aquaporin cDNAs. VvTIP2;1 isoforms had different 3′ UTRs, immediately upstream of the poly(A) tail, suggesting the presence of multiple cleavage sites for polyadenylation. Using published genome sequences of grapevine, we conducted a phylogenetic analysis of the MIPs with previously characterised MIPs from Arabidopsis. We identified 23 full-length MIP genes from the V. vinifera genome sequence of a near homozygous line (PN40024) that cluster into the four main subfamilies (and subgroups within) identified in other species. However, based on the identification of PIP2 genes in Cabernet Sauvignon that were not present in the PN40024 genome, there are likely to be more than 23 MIP genes in other heterozygous grapevine cultivars. Water transport capacity was determined for several PIPs and TIPs, by expression in Xenopus oocytes. Only VvPIP2 and VvTIP proteins function as water channels with the exception of VvPIP2;5. VvPIP2;5 differs from the water conducting VvPIP2;1 by the substitution of two highly conserved amino acids in Loop B (G97S, G100W), which was shown by homology modelling to likely form a hydrophobic block of the water pore.
Additional keywords: VvPIP1, VvPIP2, VvTIP, VvNIP, VvSIP, VvXIP.
Introduction
Grapevine is one of the worlds’ most economically important fruit crops. It is the first fruit crop to have its genome fully sequenced (Jaillon et al. 2007; Velasco et al. 2007), and now has the potential to be a model organism for future studies of fruit trees (Troggio et al. 2008). Although grapevines are able to survive in a range of soil moisture conditions, their growth and yield is determined by their total water use (McCarthy et al. 2001), so it is important to identify and characterise the molecular components of water transport. The discovery and characterisation in plants of membrane intrinsic proteins (MIPs), some of which function as water permeable pores (aquaporins), has shown that aquaporins can have primary roles in regulating plant water transport (Maurel et al. 2008). In grapevine, there are relatively few studies characterising the MIPs in general, and identifiying the roles of the MIPs in water transport (Baiges et al. 2001; Picaud et al. 2003; Vandeleur et al. 2009).
The MIP superfamily can be divided into four subfamilies; the plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin-like intrinsic proteins (NIPs) and the small basic intrinsic proteins (SIPs) (Johanson et al. 2001; Zardoya and Villalba 2001; Quigley et al. 2002; Forrest and Bhave 2008). Two new subfamilies have recently been proposed in the moss Physcomitrella patens (Hedw.) B.S.G.; the GlypF-like intrinsic proteins (GIPs) (Gustavsson et al. 2005) and the hybrid intrinsic proteins (HIPs) (Danielson and Johanson 2008). A third new subfamily called the x intrinsic proteins (XIPs) has recently been identified, with members in several dicotyledonous plants including tomato (Lycopersicon esculentum L.; Sade et al. 2009) and grapevine (Danielson and Johanson 2008). The coding region of MIPs is typically between 250 and 300 amino acid residues in length and they have a molecular mass between 26 and 34 kDa. Although the naming of the PIP and TIP groups imply a membrane location there are reports of their occurrence at different locations (Barkla et al. 1999; Alexandersson et al. 2004; Whiteman et al. 2008).
The divergence of plant MIPs is believed to have occurred early, with findings that MIPs in the moss, P. patens, fall into the same subfamilies as in the angiosperms (Borstlap 2002). MIP genes have been identified in over 30 plant species of both monocots and dicots in which they constitute a large gene family; 35 members identified in Arabidopsis (Johanson et al. 2001), 31 in maize (Zea mays L.; Chaumont et al. 2001), at least 35 in wheat (Triticum aestivum L.; Forrest and Bhave 2008) and 33 in rice (Oryza sativa L.; Sakurai et al. 2005). MIPs have been identified in woody plants such as grapevine (Baiges et al. 2001; Picaud et al. 2003; Fouquet et al. 2008; Vandeleur et al. 2009), walnut (Juglans regia L.) (Sakr et al. 2003) and olive (Olea europaea L.) (Secchi et al. 2007).
MIPs have a highly conserved structure, consisting of tandem repeats of three membrane spanning α-helical domains. Each tandem repeat has the highly conserved asparagine-proline-alanine (NPA) motif that forms the aqueous pore (Fu et al. 2000). Loops B (cytoplasmic) and E (extracellular) form short hydrophobic helices that correspond to two hemipores, dipping into the membrane to form a single transmembrane aqueous pathway. Each monomer functions as a single water pore, although aquaporins are known to form both homotetramers (Engel et al. 2000) and heterotetramers in the membrane (Neely et al. 1999; Harvengt et al. 2000). The atomic structures of several mammalian aquaporins selective for water have been determined, including AQP1 (Murata et al. 2000), AQP0 (Gonen et al. 2004; Harries et al. 2004) and AQPZ from Escherichia coli (Savage et al. 2003). A spinach plasma membrane intrinsic protein, SoPIP2;1, has been crystallised in both the open and closed conformation yielding new insights into the structural characteristics of plant aquaporins (Törnroth-Horsefield et al. 2006).
The transport selectivity of plant aquaporins has been characterised by expression in both Xenopus laevis oocytes and in yeast, and by reconstitution in proteoliposomes (Maurel et al. 2008). MIPs have a broad range of transport selectivity with various members permeable to water (Biela et al. 1999; Santoni et al. 2000; Tyerman et al. 2002), glycerol (Biela et al. 1999; Weig and Jakob 2000; Moshelion et al. 2002), urea (Gerbeau et al. 1999), CO2 (Uehlein et al. 2003), NH3 (Jahn et al. 2004; Loque et al. 2005), silicon (Ma et al. 2006) boron (Takano et al. 2006; Tanaka et al. 2008) and arsenite (Kamiya et al. 2009). The PIP2 members have been shown to have high water permeability, though there are exceptions (Zhou et al. 2007), and PIP1s have low water permeability or no apparent permeability. In maize, all ZmPIP2 proteins analysed show high water permeability and ZmPIP1a and ZmPIP1b low water permeability (Chaumont et al. 2000). The reason for this difference remains unclear, but these inactive proteins were expressed in the plasma membrane of oocytes, indicating correct targeting to the plasma membrane (Chaumont et al. 2000). Based on functional data in Xenopus oocytes, several authors have proposed that PIP1 and PIP2 isoforms interact to form heterotetramers (Fetter et al. 2004; Mahdieh et al. 2008). Such an interaction has been observed when two grapevine isoforms, VvPIP1;1 and VvPIP2;2, are co-expressed in Xenopus oocytes (Vandeleur et al. 2009), but was not observed for PIP1 PIP2 combinations from common bean (Phaseolus vulgaris L.) (Zhou et al. 2007). Direct interaction of the two subgroups has been shown using FRET imaging in living maize cells overexpressing both these isoforms (Zelazny et al. 2007). High water channel activity has also been observed in some PIP1 members including NtAQP1 from tobacco (Biela et al. 1999), BoPIP1b and BoPIP1;2 from Brassica (Marin-Olivier et al. 2000) and the Arabidopsis aquaporins, AtPIP1a, AtPIP1b and AtPIP1c (Kammerloher et al. 1994). Two PIP1 aquaporins from grape berry show low water permeability but one (VvPIP1a) has been shown to transport glycerol (Picaud et al. 2003).
The aim of the present work was to identify and functionally characterise PIP and TIP aquaporins present in the grapevine, V. vinifera cv. Cabernet Sauvignon. During this study, two high quality draft genome sequences of grapevine were reported; a near-homozygous line PN40024, bred from V. vinifera cv. Pinot Noir (Jaillon et al. 2007), and a heterozygous grapevine variety V. vinifera cv. Pinot Noir (clone ENTAV 115) (Velasco et al. 2007). Here, we present a complete analysis of the MIP superfamily in grapevine, based on sequences obtained from the sequencing of the grapevine genome, and cDNAs independently obtained in this study from Cabernet Sauvignon. The results show a large diverse family of AQPs comprising four subgroups. Functional characterisation in Xenopus laevis oocytes of some of the aquaporins identified from Cabernet Sauvignon is also presented.
Materials and methods
Plant material
One-year-old Vitis vinifera L. cv. Cabernet Sauvignon (clone LC14) rootlings (Yalumba Nursery, Barossa Valley, SA, Australia), were planted in 12 inch pots containing a modified UC soil mix (61.5% (v/v) sand, 38.5% (v/v) peat moss, 0.50 g L–1 calcium hydroxide, 0.90 g L–1 calcium carbonate, 100 g L–1 Nitrophoska (12 : 5 : 14 N : P : K plus trace elements; Incitec Pivot, Melbourne, Vic., Australia) at pH 6.8) and fertilised with 0.08 g L–1 per month of Osmocote Standard (Scotts Australia Pty Ltd, Baulkham Hills, NSW, Australia). Plants were grown in controlled temperature glasshouses maintained at 25°C day/20°C night with extended light period provided by mercury halide lamps (14 h day/10 h night). Plants were watered to field capacity every 2 days. Plant material was collected from the apical five nodes between 1000 and 1200 hours, frozen immediately in liquid nitrogen and stored at −80°C for RNA extraction.
Total and poly(A)+ RNA isolation
Total RNA was extracted from 200 to 500 mg of young stem, leaf, tendril, petiole and root tissue using a 5 M sodium perchlorate extraction buffer (5 M NaCl04, 0.2 M Tris–HCl pH 8.3, 2% (w/v) SDS, 8.5% (w/v) polyvinylpolypyrrolidone (PVPP), 1% (v/v) β-mercaptoethanol). A modified protocol of the RNeasy Extraction Kit (Qiagen, Melbourne, Vic., Australia) was used to purify total RNA (Franks et al. 2006). Poly(A)+ RNA was purified using Oligo (dT)-cellulose spun column chromatography (GE Healthcare, Piscataway, NJ, USA). The quality and purity of mRNA was checked on a denaturing agarose gel (1.2% (w/v) agarose, 1× MOPS, 8% (v/v) formaldehyde) before construction of the cDNA library and by measuring the absorbance at 260/280 nm.
Cloning of VvPIP2;1 and VvTIP1;1 by reverse transcriptase (RT)-PCR
One-step RT–PCR (Qiagen) was used to amplify VvPIP2;1 and VvTIP1;1 cDNAs from total RNA isolated from Cabernet Sauvignon petiole tissue. RNA was treated with DNase 1 (Ambion, Austin, TX, USA) before RT–PCR. PCR primers for VitisPIP2;1 and VitisTIP3 (see Table S1 available as an Accessory Publication to this paper), were used to amplify products using the following cycling conditions: 50°C for 30 min, 95°C for 15 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by cDNA extension at 72°C for 10 min. PCR products were gel purified (QIAXII Gel Purification Kit, Qiagen), cloned into pGEMT-easy (Promega, Alexandria, NSW, Australia) and transformed into E. coli XL1-B-cells. Transformants were selected using blue white colour selection and plasmid DNA purified (genelute plasmid purification kit: Sigma Aldrich, Castle Hill, NSW, Australia). All cDNA inserts were sequenced using Dye Terminator 3 (Applied Biosystems, Foster City, CA, USA) and analysed by the Institute of Medical and Veterinary Sciences (Adelaide, SA, Australia).
Complementary DNA library construction
A grapevine cDNA library was constructed from a combined mRNA pool consisting of individual mRNA extractions from stems, leaves, tendrils, petioles and roots using the Cloneminer cDNA Library Construction Kit (Invitrogen, Melbourne, Vic., Australia). The titre of the cDNA library in E.coli was determined to be 2.1 × 107 colony forming units mL–1.
Macroarray synthesis
Individual E. coli transformants were spotted into 384 well microtitre plates containing LB (supplemented with 7.5% glycerol, 50 μg mL–1 kanamycin) using a VersArray Colony Picker and Arrayer System (Biorad, Hercules, CA, USA). Microtitre plates were sealed in plastic bags, cells grown at 37°C for between 20 and 24 h, covered with aluminium seal (AlumaSeal II, Excel Scientific, CA, USA) and stored at –80°C. Hybond N+ nylon membranes (GE Healthcare) were placed onto large plastic plates, spotted with six individual 384 well plates containing cloned grapevine cDNAs. Membranes were placed colony side up, onto large plates containing solid LB media (supplemented with 50 μg mL–1 kanamycin) and grown overnight at 37°C. Cells were lysed, DNA denatured and fixed to the membrane following the protocol of Sambrook and Russel (2001). Membranes were air-dried for 30 min and DNA fixed using a UV transilluminator (GE Healthcare, Piscataway, NJ, USA).
Screening of cDNA library
Forward and reverse PCR primers (Sigma Aldrich) were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi, accessed October 2003) to the 5′ and 3′ end of Vitis Rootstock Richter 110 aquaporin cDNAs and V. vinifera Pinor Noir AQP cDNAs (Table S1).
Using the appropriate PCR primers, cDNA fragments were amplified from purified plasmid cDNA library template with the Digoxygenin (DIG)-labelling PCR kit (Roche Diagnostics, Castle Hill, NSW, Australia) and TaqTi polymerase (Fischer Biotech, Adelaide, SA, Australia). PCR cycling conditions were as follows: 95°C for 15 min for 1 cycle; 95°C for 30 s, 60°C for 30 s, 72°C for 1.5 min for 35 cycles; extension at 72°C for 7 min. DIG-labelled PCR products were separated by gel electrophoresis, gel purified and cloned into pGEMT-Easy (Promega). Inserts were sequenced from both the 3′ and 5′ end to verify the amplified product. Equal amounts of each DIG-labelled cDNA probe were combined and used together to screen the complete cDNA library under high stringency conditions. Spotted cDNA membranes were prehybridised in DIG-Easy Hyb solution for 1 h at 42°C. The combined probe set was diluted in DIG-Easy Hyb solution, denatured at 100°C for 5 min and then cooled quickly on ice. Denatured probe was added to the membranes and hybridised at 42°C overnight in a roller tube. Membranes were washed twice each in the following solutions: low stringency wash buffer (2 × SSC, 0.1% SDS) at room temperature for 5 min; high stringency wash buffer (0.1 × SSC, 0.1% SDS) at 68°C for 15 min; and 2 × SSC. DIG-labelled DNA was detected by chemiluminescence using an anti DIG-alkaline phosphatase antibody and CDP star (Roche Diagnostics). Positive colonies were detected using the Molecular Imager ChemiDoc XRS System (Biorad) and ImaGene 6.0 software (BioDiscovery, El Segundo, CA, USA) was used to locate and quantitate these signals on the macroarray.
Rapid amplification of cDNA ends
Rapid amplification of cDNA ends (RACE-PCR) was performed to amplify the 5′ end of two incomplete aquaporin cDNAs. Primers were designed to amplify the 5′ end of the partial length aquaporin cDNAs, VvPIP2;3 and VvPIP1;4, using 5′ RACE-PCR (Table S1). The forward primer was designed in the multiple cloning site of pDONR222 (pDONR222F) and specific reverse primers were designed to the 5′ end of each partial length cDNA (VvPIP1;4R and VvPIP2;3R). 5′ RACE-PCR was performed on the cDNA library template using high fidelity platinum Taq polymerase (Invitrogen). PCR cycle for isolate VvPIP2;3 and VvPIP1;4 was as follows: 1 cycle of 95°C for 2 min; 35 cycles of 95°C for 30 s, 60.3°C for 30 s, 68°C for 1 min; 1 cycle of 68°C for 7 for min. PCR products were separated by gel electrophoresis, gel purified, cloned into pGEMT-Easy and transformed into E. coli XL1-B. Positives colonies were selected, plasmid DNA purified and then sequenced from both 5′ and 3′ ends.
pGEMHE-DEST plasmid construction
The oocyte expression vector, pGEMHE (Liman et al. 1992) that contains the 5′ and 3′ β-globin untranslated regions flanking the polylinker (Krieg and Melton 1984), was converted to a Gateway destination vector using the gateway vector conversion system (Invitrogen). pGEMHE was linearised with BamHI, end-filled using Klenow (Roche) and dephosphorylated with antartic phosphotase (NEB, Ipswich, MA, USA). The reading frame cassette A (RfA) was blunt end ligated to the linearised pGEMHE vector and subsequently transformed into E. coli strain DB3.1. (Invitrogen). The reading frame cassette A contains the chloramphenical resistance gene (CmR) and the ccdB gene flanked by the attR1 and attR2 sites. Plasmid preparations of putative pGEMHE/gateway transformants resistant to antibiotic (chloramphenical, ampicillin) selection were digested with Bsr G1 (NEB) and sequenced with Gateway forward and reverse primers to confirm conversion to a gateway vector (Table S1). The resulting plasmid was designated pGEMHE-DEST (Fig. S1 available as an Accessory Publication to this paper).
Amplification of attB PCR products
AttB PCR primers were designed according to the Gateway Technology Manual (Invitrogen) to amplify attB PCR products of VvPIP2;1 and VvTIP1;1 (Table S1). Pwo DNA Polymerase (Roche) was used to amplify attB/VvTIP1;1 by PCR from the cDNA library while AttB/VvPIP2;1 was amplified from a previously cloned VvPIP2;1 cDNA. The PCR cycling conditions were as follows: 94°C for 2 min; 94°C for 15 s, 55°C for 30 s, 72°C for 1 min cycled 10 times; 94°C for 15 s, 55°C for 30 s, 72°C for 1 min plus 5 s each cycle for 15 cycles; and extension at 72°C for 7 min. The attB PCR products were PEG purified (according to Gateway Technology Manual) and recombined into pDONR222 using BP Clonase (Invitrogen). The Gateway BP reaction facilitated recombination of the attB PCR products into pDONR222 to create attL entry clones of VvPIP2;1 and VvTIP1;1. LR Clonase was used to recombine VvPIP2;1 and VvTIP1;1 into pGEMHE-DEST. Entry clones were digested and sequenced to confirm correct PCR product.
Cloning of AQP cDNAs into oocyte expression vector
Full-length V. vinifera aquaporin cDNAs identified in the library screen were recombined into the pGEMHE-DEST vector using the LR recombination reaction (Invitrogen). The cDNA of HsAQP1 from human red blood cells was obtained from Daniel M. Roberts (University of Tennessee, Knoxville, TN, USA). HsAQP1 cDNA was cloned into the vector XβG-ev1 between the 5′ and 3′ untranslated region (UTR) of the Xenopus β-globin cDNA (Preston et al. 1992). For cRNA synthesis, pGEMHE-DEST based vectors were digested with Nhe 1 at 37°C overnight to linearise the plasmid, while HsAQP1 XβG-ev1 plasmid was digested with Bam H1. Complementary RNA was transcribed using 1 μg of linearised DNA with the mMESSAGE mMachine Kit (Ambion) utilising the T7 promoter of pGEMHE, according to the manufacturers instructions. Phenol : chloroform : isoamyl alcohol (25 : 24 : 1 v/v) (Sigma Aldrich) was used for extraction and isopropanol for precipitation of the synthesised cRNA. cRNA was quantitated and checked for purity using a spectrophotometer (Smartspec; Biorad) and by size separating on a denaturing RNA gel.
Harvesting oocytes
Oocytes were harvested from Xenopus laevis using the protocol described by Hill et al. (2005). Following harvest, oocytes were washed twice in cold Ca-free Ringers solution (96 mM NaCl, 2 mM KCl, 5 mM MgCl2; 5 mM HEPES, pH 7.6), then defolliculated in 1.7% (w/v) collagenase in Ca-free Ringers solution for 90 min with rotation. Oocytes were washed five times in Ca-Ringers solution (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, 6 mM CaCl2, 5 mM HEPES, pH 7.6) to remove all traces of collagenase. Oocytes were transferred to Ca-Ringers solution supplemented with antibiotics (100 μg mL–1 tetracycline-HCl, 100 units mL–1 penicillin, 100 μg mL–1 streptomycin) and kept at 18°C overnight. Defolliculated oocytes were microinjected with the Nanoject II Auto-Nanolitre Injector (Drummond Scientific, Broomall, PA, USA) into the cytoplasm with 50 ng of capped cRNA or with DEPC treated water (control) and incubated for three days at 18°C in Ca-Ringers solution supplemented with antibiotics 100 μg mL–1 tetracycline-HCl, 100 units mL–1 penicillin and 100 μg mL–1 streptomycin.
Oocyte swelling and acidification assay
Oocytes were preincubated at room temperature in an iso-osmotic solution (Ca-Ringers solution) for 5 min, 64–72 h after cRNA injection, and then transferred to a hypoosmotic solution (Ca-Ringers solution diluted 5-fold with sterile water) at which time swelling was measured for 2 min. Oocytes were viewed with a Nikon SMZ800 light microscope with Vikam colour camera at 2× magnification and imaged with Global Laboratory Image/2 software (Data Translation, Marlboro, MA, USA). Images were acquired every 4 s for 2 min. Osmotic permeability (Pf ) was calculated for water injected and cRNA injected oocytes from the initial rate of change in relative volume (dV rel/dt) i calculated from projected images assuming the oocytes were spherical:

where Vi and Ai are the initial volume and area, respectively, Vw is the partial molar volume of water; and ΔCo is the change in external osmolarity. At least two independent experiments with 5–8 oocytes each were conducted for each gene. Osmolarity of each solution was determined using a vapour pressure osmometer 5500 (Wescor, Logan, UT, USA). Cytosolic pH was lowered using an external solution of 50 mM Na-acetate at pH 5 in Ca-Ringers solution (Tournaire-Roux et al. 2003). Controls contained 50 mM Na acetate at pH 7. Swelling assays were conducted as above with oocytes incubated in 50 mM Na-acetate pH 5 solution for 5 min to allow acidification of the oocyte cytosol. Oocytes were then transferred to a hypotonic solution (five times diluted with MilliQ water) and swelling imaged for 2 min.
Modelling of AQP
Homology modelling was used to generate a predicted structure for the VvPIP2;5 protein. The model was generated using SWISS-MODEL (Schwede et al. 2003) with the structure of the open conformation of SoPIP2;1 (PDB number 285F) selected as a template. Pictures were generated with Deep View-Swiss PDB viewer.
Sequence analysis
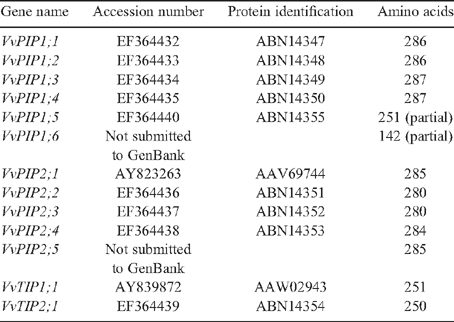
Sequences identified in the library screen were analysed using the BLASTn (nucleotide) and tBLASTn (translated nucleotide) algorithms at National Centre for Biotechnology Information (Altschul et al. 1997). MacVector 9.0 (Oxford Molecular Ltd, Oxford, UK) was used to translate sequences and ClustalW was used to generate multiple sequence alignments using blosum30 matrix and slow mode. The open gap penalty and extended gap penalty was set at 10.0 and 0.05, respectively. Phylogenetic analysis was performed using a neighbour joining analysis of the deduced amino acid sequences (bootstrap 100 repetitions) using MEGALIGN 4.0. All novel aquaporin sequences identified were submitted to GenBank (accession numbers shown in Table 1).
|
Results
Cloning of AQP cDNAs
A total of ~30 000 E. coli transformants containing grapevine cDNAs were screened under high stringency conditions for PIP and TIP cDNAs. Thirty colonies were identified based on initial DIG hybridisation with the combined DIG-labelled aquaporin probe set. Each positive colony was regrown in liquid culture, plasmid purified and digested to release the cDNA insert. Plasmids containing cDNAs greater than ~300 bp were sequenced. Sixteen cDNAs were positively identified as being members of the MIP super family by BLASTn searches using GenBank. Of these eight were found to be full-length cDNAs and eight were identified as partial length cDNAs. The cDNAs ranged in size from 278 bp to 1216 bp. The sequences were compared with previously identified MIP genes from both grapevine and Arabidopsis thaliana and were annotated according to the nomenclature set out by Johanson et al. (2001). Multiple cDNA isolates, encoding VvPIP1;2, VvPIP2;4 and VvTIP2;1, were identified in the library screen indicating degeneracy in the library and potentially high expression of these genes within the plant. Of the cDNAs identified, eleven were found to be novel grapevine aquaporins. Two aquaporin cDNAs not isolated in the screen VvPIP1;1 and VvPIP1;3, were identified from sequencing of PCR generated probes. These have been included in the bioinformatics analysis, but were not functionally characterised in Xenopus oocytes.
Full-length sequences were obtained for two partial cDNAs using 5′ RACE-PCR. Primers designed to the multiple cloning site of pDONR222 and the 5′ end of VvPIP2;3 and VvPIP1;4 partial clones successfully amplified major products from the cDNA library at the expected size of 500 bp and 600 bp, respectively. These fragments were sequenced and aligned to the corresponding partial cDNA, primers designed to the 5′ and 3′ end of the full-length fragment and subsequently amplified with PCR. The full-length cDNA of VvPIP2;3 is 1225 bp, encoding a polypeptide of 280 amino acids, while the cDNA of VvPIP1;4 is 1061 bp, encoding a PIP1 aquaporin, 287 amino acids in length. In total, 13 putative aquaporin PIP and TIP cDNAs were obtained (Table 1). Eleven cDNAs were submitted to GenBank and designated an accession number (Table 1).
Bioinformatics
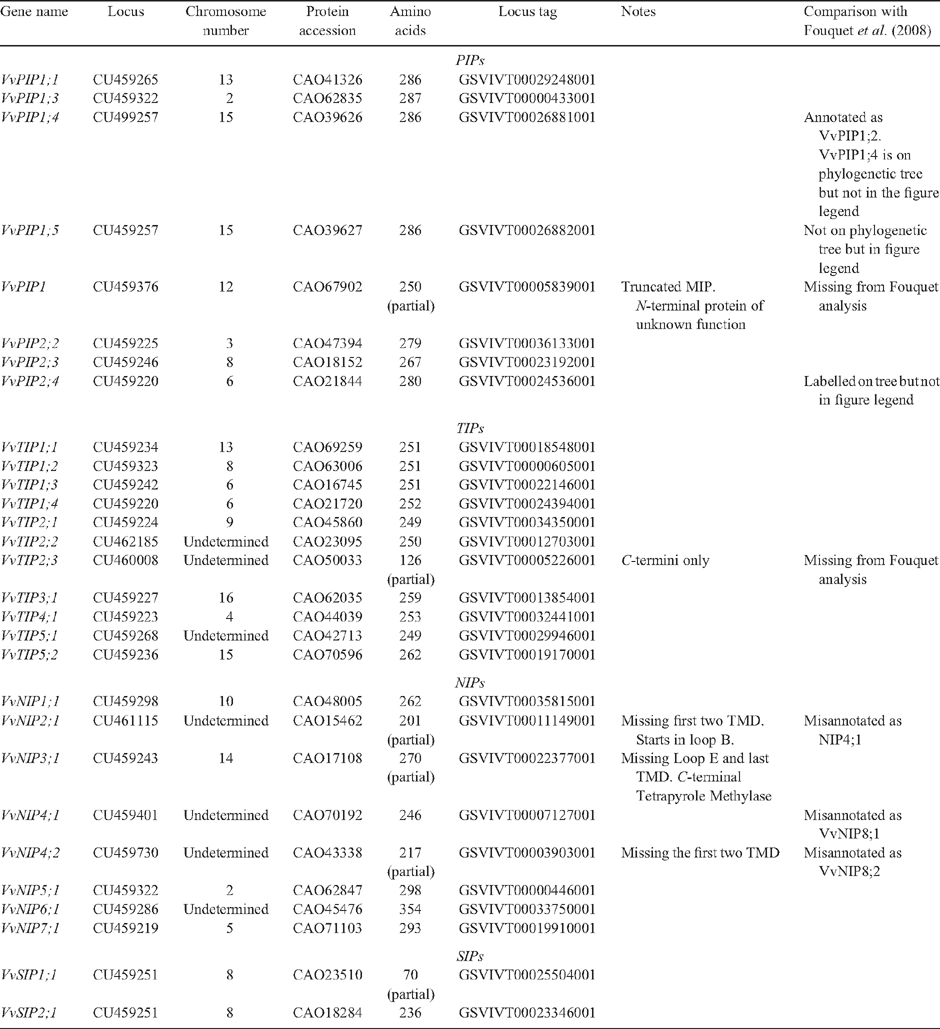
The completion of two draft sequences of the V. vinifera genome, clone PN40024 (Jaillon et al. 2007) and Pinot Noir ENTAV 115 (Velasco et al. 2007), has made it possible to identify putative MIP genes in the grapevine genome. Currently, there are 83 grapevine related MIPs annotated in GenBank, 73 from the wine grape V. vinifera (cvv. Syrah, Cabernet Sauvignon, Pinot Noir and Nebbelio), eight from the rootstock Richter-110 (Vitis berlandieri × Vitis rupestris) and one from the Chinese wild grape (Vitis pseudoreticulata). By using BLAST searches in GenBank, we identified 29 putative MIP genes in grapevine (clone PN40024): 23 of these encoded full-length proteins and six encoded partial MIP proteins (Table 2). Nineteen putative aquaporin genes (15 of these are full-length) were also identified in the heterozygous Pinot Noir genome (Table S2), including a putative gene encoding a XIP protein (Danielson and Johanson 2008). Several genes encode partial proteins, missing one or more transmembrane domain, indicating these may be non-functional pseudogenes, or misannotated from the genome sequencing project.
|
All full-length PIP2 cDNAs identified in Cabernet Sauvignon are ~1200 bp in length and when compared at the nucleotide level show between 69 and 99% homology. We observed VvPIP2;4 and VvPIP2;5 are 99% homologous at the nucleotide level, and VvPIP2;1, VvPIP2;4 and VvPIP2;5 are 99% homologous at the amino acid level. VvPIP2;1 and VvPIP2;4 differ by a conservative amino acid substitution at position 193 from arginine to lysine. Similarly, VvPIP2;1 and VvPIP2;5 were found to be highly similar with only two amino acid substitutions, Gly 97 Ser and Gly 100 Trp, both of which are located in the highly conserved loop B. VvPIP2;3 and VvPIP2;2 are 77 and 80% homologous to VvPIP2;1, respectively. The PIP1 cDNAs are ~1100 bp in length and between 68 and 99% homologous. VvPIP1;2 and VvPIP1;4 are 99% homologous at both the nucleotide and at the amino acid level with only two amino acid differences in the coding region, Ile 247 Val and Arg 250 Lys.
Five cDNA isolates from Cabernet Sauvignon were found to have identical nucleotide sequences in their coding region. The deduced amino acid sequence of these cDNAs encode the protein, VvTIP2;1. Interestingly, the only difference observed between the isolates was in the 3′ untranslated region. Two isolates were both found to have a 15 bp deletion in the 3′ UTR, immediately upstream of the poly(A) tail, compared with the other three isolates. Analysis of other Vitis TIP2;1 homologues on the database, show that several these also have deletions in the 3′ UTR immediately upstream of the poly(A) tail suggesting the presence of different cleavage sites for polyadenylation. VvTIP2;1 is 72% identical to VvTIP1;1 at the nucleotide level and 72% at the amino acid level. Multiple cDNA isolates of VvPIP1;2 and VvPIP2;4 were also identified in the library screen, however, no sequence differences were observed in their 3′-UTR.
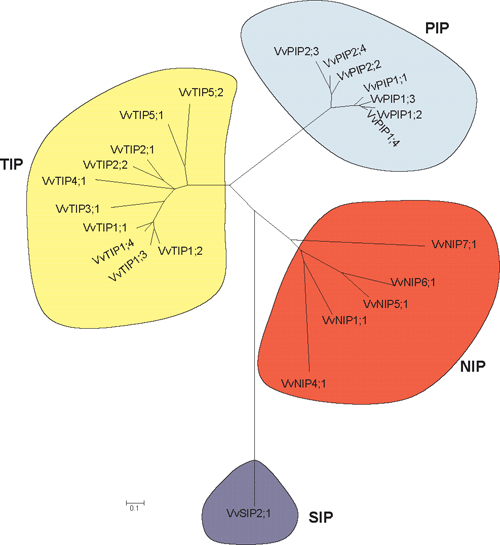
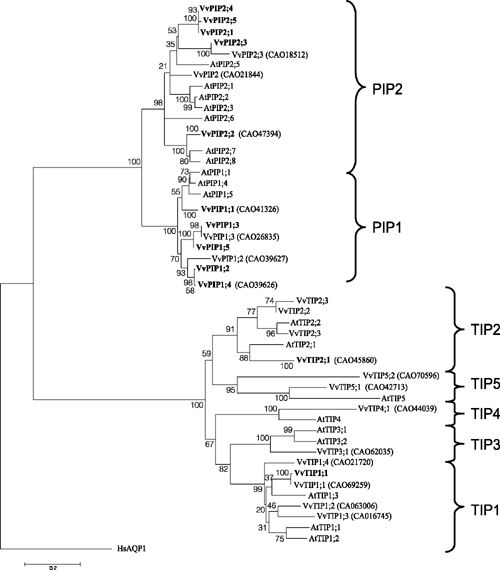
To correctly annotate the grapevine MIPs, multiple alignments of translated sequences were generated with ClustalW and compared with Arabidopsis MIPs. Evolutionary history was inferred using the neighbour-joining method. These were subsequently annotated according to the nomenclature proposed by Johanson et al. (2001). The full-length grapevine MIPs range in size from 236 to 354 amino acids (Table 2). All grapevine MIPs contain the conserved NPA motif in loops B and E, with the exception that in several of the NIPs, the alanine can be replaced by a Ser or Val (Table S3). In VvSIP2 the alanine in loop B is replaced by leucine. The 23 grapevine MIPs cluster into the four distinct MIP subfamilies: PIPs, TIPs, NIPs and SIPs in a similar fashion to that of Arabidopsis (Figs 1, 2). The PIP members show divergence into the two subgroups, PIP1 and PIP2, and the grapevine TIPs cluster into five groups, TIP1, TIP2, TIP3, TIP4 and TIP5 (Fig. 2). Several grapevine aquaporins cluster on one branch (VvPIP2;1, VvPIP2;4 and VvPIP2;5) separate to those identified in Arabidopsis which form separate branches, respectively. This multiplicity of highly related aquaporins has been seen previously for both monocots (Chaumont et al. 2001; Sakurai et al. 2005) and dicots (Johanson et al. 2001).
|
|
Structural characteristics and conserved motifs
All MIP members identified in V. vinifera cv. Cabernet Sauvignon show characteristic sequence motifs found throughout the MIP superfamily. Multiple sequence alignments of the deduced amino acid sequences, performed using ClustalW, show the high homology between the PIP and TIP members identified in this study (Fig. 3). The PIP1 members show an extended N-terminal tail of 14 amino acids compared with the PIP2 members. Conversely, the PIP2 members have an extended C-terminal tail comprising eight amino acids. TIP members have a much shorter N-terminal tail than all the PIPs. Several residues are completely conserved in all grapevine PIP and TIP cDNAs identified, with particularly high conservation found in the functional loops B and E. Several of these residues are also conserved in loops B and E of mammalian aquaporins (Murata et al. 2000). Hydropathy analysis of VvPIP1;1, VvPIP2;1 and VvTIP1;1 identified six transmembrane α spanning domains that are separated by hydrophilic loops (data not shown). The C- and N termini were predicted to be cytoplasmic and the highly conserved hydrophobic NPA motifs were in the loop between TMD1 and TMD2, designated loop B, and loop E between TMD5 and TMD6.
Protein similarity between Vitis species and cultivars is greater than 98% for all proteins identified (with the exception of the PIP2;3 homologue from PN40024) indicating early divergence of the PIP and TIP genes within the genus Vitis. Closely related proteins (with homology >98%) have most likely arisen from recent gene duplications, as has been reported in other species such as maize (Chaumont et al. 2001). The PIP2;3 homologue from PN40024 appears to be missing 20–30 amino acids and therefore may be annotated incorrectly.
Functional analysis
The functionality of selected full-length V. vinifera AQP cDNAs was assessed by expression in Xenopus oocytes. Swelling was measured in response to bathing in hypo-osmotic solution, ~3 days after injection with cRNA. The water permeability (Pf ) was calculated and compared with the water-injected oocytes (control) (Fig. 4a ). HsAQP1, from human red cells, was used as a positive control. The mean Pf value for AQP1 was 22 ± 4 × 10−3 cm s–1. The mean Pf of VvPIP2 proteins ranged from between 8.0 and 27.0 × 10−3 cm s–1. The Pf value for VvPIP2;5 was 0.6 × 10−3 cm s–1, higher than the water injected control 0.1 × 10−3 cm s–1 (although not significant P > 0.05). PIP1 proteins (VvPIP1;2 and VvPIP1;4) also showed low permeability with no significant difference compared with the water injected controls, 0.5 × 10−3 cm s–1. The Pf value for VvTIP2;1 was 9.0 ± 5.0 × 10−3 cm s–1, but VvTIP1;1 was significantly lower at 5.0 ± 2.0 × 10−3 cm s–1.
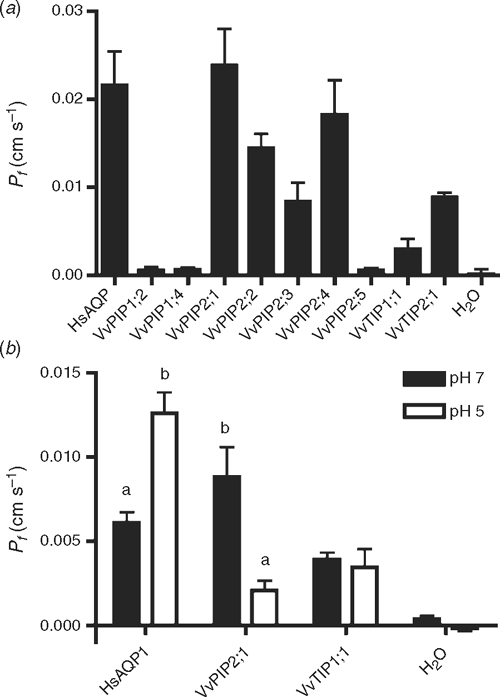
|
A highly conserved His residue in loop D (His197) has been implicated in channel gating under anoxic conditions in an Arabidopsis PIP2 aquaporin (Tournaire-Roux et al. 2003). The corresponding His residue (His196) is conserved in all VvPIP aquaporins identified. In order to check the effect of acidic cytosolic pH on regulation of selected Vitis aquaporins, oocytes expressing HsAQP1, VvPIP2;1, VvTIP1;1 and H2O injected controls, were acid stressed with a weak acid at low pH. A significant decrease in Pf from 9.0 ± 1.8 × 10−3 to 2.0 ± 1.0 × 10−3 was observed for VvPIP2;1 when exposed to Na-acetate pH 5, compared with Na-acetate pH 7 (P < 0.001). The Pf value of VvTIP1;1 was unaltered. HsAQP1 showed a 2-fold increase in Pf when exposed to Na-acetate pH 5 solution, compared with the pH 7 control solution (P < 0.001).
Discussion
Identification and phylogenetic analysis of grapevine aquaporins
The screening of a V. vinifera cv. Cabernet Sauvignon cDNA library for PIP and TIP cDNAs has resulted in the identification of 13 cDNAs, 11 of these encode full-length proteins and two encode partial proteins. These were identified as members of the MIP superfamily based on homology to other plant aquaporins. Of these cDNAs, five are PIP2 aquaporins, six are PIP1 and two are TIP aquaporins. Using the high quality draft genomic sequence of the homozygous V. vinifera Pinot Noir clone PN40024 (Jaillon et al. 2007), we identified 23 full-length genes encoding MIP proteins and have designated these into their respective subgroups: the PIPs, TIPs, NIPs and SIPs. Six partial sequences were also identified in the grapevine genome, one PIP, one TIP, three NIPs and one SIP. Based on our bioinformatics and phylogenetic analysis, we have proposed different nomenclature for several the PN40024 genes (Table 2) compared with a previous study (Fouquet et al. 2008). Furthermore, Danielson and Johanson (2008) recently reported a gene encoding a XIP protein in the genome sequence of Pinot Noir (ENTAV 115). No members of the XIP family have been found in Arabidopsis (Danielson and Johanson 2008), or in the grapevine genome of PN40024.
Using the genomic sequence as a reference we believe we have isolated the majority of PIP genes found in Cabernet Sauvignon. The PIPs identified in this study are highly homologous at both the nucleotide and amino acid level. This is characteristic of PIP aquaporins with very high homologies found in other species such as Arabidopsis (Johanson et al. 2001; Quigley et al. 2002), Brassica (Marin-Olivier et al. 2000), walnut (Sakr et al. 2003), maize (Chaumont et al. 2001), rice (Sakurai et al. 2005) and wheat (Forrest and Bhave 2008).
A phylogenetic analysis of the PIPs from the monocot maize and the dicot Arabidopsis, indicate that the divergence of these aquaporin genes occurred after the monocot – dicot divergence (Chaumont et al. 2001). Comparison of the V. vinifera aquaporins with Arabidopsis aquaporins clearly shows that the divergence of plant aquaporins into the four subfamilies occurred early in evolution (Danielson and Johanson 2008). The grapevine aquaporins segregate onto distinct branches within each subfamily indicating that highly homologous genes most likely arose from recent gene duplications. An example of this is the high homology that exists between the cDNAs for VvPIP2;1, VvPIP2;4 and VvPIP2;5. Note that VvPIP2;1 and VvPIP2;5 are missing from the PN40024 sequence (Fig. 1) but VvPIP2;4 is present and may perform the same function as the missing genes. This high nucleotide homology has been seen for aquaporins in other species such as maize, where ZmPIP1–3 and ZmPIP1–4 have 98% nucleotide homology and encode identical proteins. In maize, these genes were also shown to have a similar transcript distribution pattern, and the authors hypothesised that the role of having two genes encoding identical proteins may be to increase the expression levels of the duplicated genes (Chaumont et al. 2001). VvPIP1;2 and VvPIP1;4 are 99% identical at the nucleotide level and 100% similar at the amino acid level. The transcript abundance and tissue location in grapevine of each of these isoforms is currently unknown.
The V. vinifera TIP aquaporins have lower homologies than observed for the PIP aquaporins. This has been seen previously in both Arabidopsis (Johanson et al. 2001) and maize TIPs (Chaumont et al. 2001). The full-length cDNA of VvTIP2;1 was independently identified in the library screen five times, indicating that this gene is most likely highly expressed throughout the plant. The Arabidopsis homologue δ-TIP (renamed AtTIP2;1), has been shown to be highly expressed in the leaves (Alexandersson et al. 2005). Further, two of the VvTIP2;1 cDNAs identified had a 15-bp deletion at the 3′ end of the cDNA, immediately before the poly(A) tail. Differences in both the 3′ non-coding region and the length of the poly(A) tail have been reported previously for Vitis PIP1b cDNAs (Picaud et al. 2003). Plant polyadenylation cleavage sites have a consensus signal which comprises a pyrimidine/adenosine dinucleotide (YA) within a U rich region (Hunt 1994). The presence of multiple cleavage sites in the 3′ UTR can result in the addition of the poly(A) tail at different positions. This may affect the stability of the transcript, translation of the protein and trafficking of the transcript to the cytoplasm, and ultimately may lead to changes in protein activity.
Five full-length grapevine NIPs were identified in the grapevine genome of V. vinifera clone PN40024. Phylogenetic analyses show the NIPs are more divergent than both the grapevine PIP and TIP aquaporins as has been reported previously for other plants (Wallace et al. 2006). The NIPs can be divided into two subgroups: group I (NIP1, NIP2, NIP3 and NIP4) and group II (NIP5, NIP6 and NIP7) based on solute permeability and the structure of selectivity-determining ar/R filter (Wallace et al. 2006). We propose that NIP4;1, NIP8;1 and NIP8;2, previously reported by Fouquet et al. (2008) should be renamed as VvNIP2;1, VvNIP4;1 and VvNIP4;2, respectively. Functional characterisation of plant NIPs have shown they are permeable to a broad range of small solutes (Wallace et al. 2006), however, to date, none of the grapevine NIPs have been functionally characterised.
The SIPs comprise the smallest subgroup and are more divergent,than the other MIPs (Chaumont et al. 2001; Johanson et al. 2001) in particular the NPA motif in loop B is not conserved and instead consists of the motif asparagine-proline-threonine (NPT) or asparagine-proline-leucine (NPL) (Chaumont et al. 2001). The SIP identified from grapevine, designated as a VvSIP2, has the motif NPL in loop B consistent with SIPs identified in other higher plants.
Water channel activity
Nine of the full-length AQP cDNAs from the grapevine V. vinifera cv. Cabernet Sauvignon have been assessed for functionality by expression in Xenopus laevis oocytes. All VvPIP2 AQPs examined (with the exception of VvPIP2;5) function as water channels with high water permeability, as has been shown previously for other plant PIP2 aquaporin members (reviewed by Tyerman et al. 2002). Water permeability varied slightly between the different V. vinifera PIP2 isoforms with VvPIP2;1 and VvPIP2;4 having the highest Pf values. These two genes are very closely related (99% homologous) with only one conservative amino acid substitution at position 209 from Arg to Lys. VvPIP2;3 and VvPIP2;2 are between 74% and 82% homologous to the other V. vinifera PIP2 aquaporins and have slightly lower Pf values. Both the VvPIP1 AQPs examined here showed low or no water transport activity, as shown previously for VvPIP1;1 (Vandeleur et al. 2009). VvPIP1;2 and VvPIP1;4 are 89% identical to the previously characterised VvPIP1b, and 98 and 99% identical to PIP1a, respectively (Picaud et al. 2003). Both of these PIP1 AQPs had no permeability for water, but PIP1a facilitated glycerol uptake (Picaud et al. 2003). Given their high homology to VvPIP1a and VvPIP1b, it is likely that VvPIP1;2 and VvPIP1;4 may also be permeable to glycerol. The inactivity of VvPIP1 aquaporins when expressed in Xenopus oocytes may also be a result of incorrect trafficking and/or insertion into the oocyte membrane. Although inactive maize PIP1 aquaporins were shown to be trafficked correctly to the plasma membrane (Chaumont et al. 2000), this would need to be confirmed for VvPIP1 members. It has also been postulated that the inactivity of PIP1 aquaporins in oocytes, may be because they require a positive regulator for functionality that is not present in oocytes (Fetter et al. 2004). Using affinity chromatography, Fetter et al. (2004) showed that co-expression of PIP1 and PIP2 aquaporins resulted in both being targeted to the oocyte membrane, possibly forming heterotetramers. A role in planta for the formation of heterotetramers has yet to be demonstrated. Mammalian AQP1 was used as a positive control for the Vitis aquaporins. Similar Pf values for AQP1 were observed to those shown previously (Pf ~20 × 10−3 cm s–1) (Preston et al. 1992).
Until recently, the molecular basis of aquaporin gating in both plants and mammals remained unknown. However, it is known that plant aquaporins are gated by pH (Tournaire-Roux et al. 2003; Fischer and Kaldenhoff 2008), calcium (Alleva et al. 2006) and phosphorylation (Johansson et al. 1996; Johansson et al. 1998; Prak et al. 2008; Endler et al. 2009). The mammalian aquaporin, AQP0, has been shown to be regulated by external pH, when expressed in Xenopus oocytes (Nemeth-Cahalan and Hall 2000; Nemeth-Cahalan et al. 2004; Hedfalk et al. 2006). The effect of cytosolic pH on gating of human AQP1, VvPIP2;1 and VvTIP1;1 was examined by expression in Xenopus oocytes (Fig. 4b ). Our results show a 2-fold increase in Pf of AQP1 at pH 5. Human AQP1 has previously been shown to be insensitive to external pH in the range 5.5 to 8.0, although both acid and alkaline pH sensitivity was induced by the addition of histidines in loops A and C (Nemeth-Cahalan et al. 2004). Although mammalian AQP1 does not contain a His residue in loop D, there is a stretch of highly charged residues in this region which may respond to the presence of cytosolic protons and result in the increase in Pf . There is also a His in the extracellular part of loop E (Nemeth-Cahalan and Hall 2000) that may account for the response at the lower external pH used in our experiments.
Tournaire-Roux et al. (2003) showed plant PIPs to be regulated by cytoplasmic pH. Mutation of a conserved His residue (His 197) in the intracellular loop D of the Arabidopsis plasma membrane aquaporin, AtPIP2, showed reduced effects in water permeability to a reduction in cytosolic pH. The corresponding His residue is conserved throughout the PIP family (but not the TIPs) and is found in all V. vinifera PIP members identified in this study. Reduction of cytosolic pH with Na-acetate pH 5, resulted in a significant decrease in water permeability of VvPIP2;1 not observed for VvTIP1;1. The X-ray structure of SoPIP2;1 in the closed confirmation supports the proposed mechanism of pH regulated gating (Törnroth-Horsefield et al. 2006). Conservation of this His residue (His 196) in VvPIP2;1, combined with the decrease in Pf observed by acidifying the cytosol, is evidence that His 196 is most likely protonated as is the case for SoPIP2;1 and AtPIP2.
Structural characteristics of grapevine aquaporins
Loop D in the PIP aquaporin members differs from other plant aquaporins, in that there are usually four to seven additional amino acid residues. Several these residues in loop D of SoPIP2;1 have been identified as being involved in gating of the channel (Törnroth-Horsefield et al. 2006). The amino acid alignment of the V. vinifera aquaporins shows that all the PIPs identified have four additional amino acid residues in loop D, not found in the TIP aquaporins. In SoPIP2;1, Leu 197 was identified as a key residue, thought to effectively form a lid to cap the channel, and in combination with other conserved residues His 99, Val 104 and Leu 108, contribute to the hydrophobic barrier that blocks the pore region. Leu 200 in VvPIP2;1, corresponding to Leu 197 in SoPIP2;1, is also fully conserved in all the PIP members, as are His 102, Val 108 and Leu 111. In the closed conformation loop D occludes the pore from the cytosol.
VvPIP2;5, identified in the cDNA library screen of Cabernet Sauvignon, has two amino acid substitutions, Gly 97 is substituted for Ser, and Gly 100 is substituted for Trp, in the highly conserved region of loop B. Both Gly residues are completely conserved in all plant PIP members. Gly 100 is highly conserved in the MIP superfamily and it is also conserved in mammalian aquaporins. The structural model of AQP1 shows this glycine faces the inside of the aqueous pore (Murata et al. 2000). This, combined with the conservation of this residue throughout the aquaporin family, indicates it most likely plays a crucial role in water transport through the channel. Ser 99 is conserved in all identified PIP, TIP and NIP aquaporins from V. vinifera and the corresponding serine residue in AQP1 forms a hydrogen bond with Tyr 137 which helps to stabilise loop B (Mitsuoka et al. 1999). The substitution of the Gly residue by a Trp residue at position 100 may also affect the stability of this interaction. Therefore, it is expected that the addition of the Trp 100 will significantly affect the structure and/or functionality of the protein. It is interesting to note this gene is not present in either of the V. vinifera genome sequences which may indicate a novel feature of the Cabernet Sauvignon genome. Semiquantitative PCR showed very low levels of expression of this gene in the vegetative tissues (results not shown). EST database searches have revealed the presence of a Trp amino acid at the same position in a PIP1 cDNA (AI443117) identified from the root tissue of soybean (Glycine max). Comon bean PvPIP2;2 also has amino acid substitutions in the highly conserved Loop B and it is also non-functional in Xenopus oocytes (Zhou et al. 2007). The functional role of these genes in planta are unknown.
An alignment of loop B with VvPIP2;1 and SoPIP2;1 is shown with the two residues highlighted (Fig. 5). A homology model of the VvPIP2;5 structure was generated using the open structure of SoPIP2;1 as a template (Fig. 5). As both Ser and Trp are larger than the Gly residues they replace, the pore mouth becomes more crowded in VvPIP2;5 and both residues make contact with other parts of the protein that are not present in the SoPIP2 structure. In particular, because Trp 100 is large and hydrophobic, it may block the pore. This could explain the very low water permeability observed for VvPIP2;5 when expressed in Xenopus oocytes. A point mutation of the corresponding residue in AQP4 (Gly 72 Trp) showed the abolition of water permeability and a decrease in trafficking of the protein to the oocyte plasma membrane (Shi and Verkman 1996). It is therefore possible that the substitution of Gly 100 Trp in VvPIP2 could interfere in correct protein folding and/or trafficking to the plasma membrane.
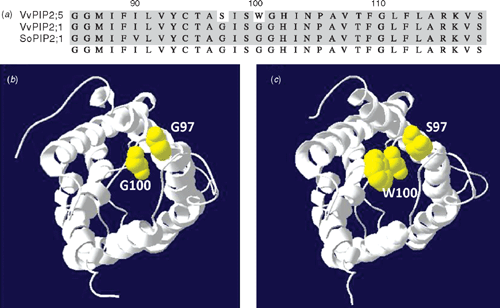
|
In summary, we have isolated grapevine PIPs and TIPs, and have shown several of these are permeable to water when expressed in Xenopus oocytes. Some have novel features that require further study. From in silico analysis of the grapevine genome sequence of a near homozygous line (PN40024), we present an alternative classification of grapevine MIPs from that already published (Fouquet et al. 2008) We identified 29 MIP genes belonging to four subfamilies, 23 of which encode full-length proteins. Further examination of grapevine MIPs is required to elucidate their functional roles in planta.
Acknowledgements
We thank Wendy Sullivan for expert technical assistance, Christa Niemietz for assistance with Xenopus oocytes and Lars Bredmose for assistance with the macroarray. This research was supported by the Australian Research Council, and the Grape and Wine Research and Development Corporation.
Alexandersson E,
Saalbach G,
Larsson C, Kjellbom P
(2004)
Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant & Cell Physiology 45, 1543–1556.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Alexandersson E,
Fraysse L,
Sjovall-Larsen S,
Gustavsson S,
Fellert M,
Karlsson M,
Johanson U, Kjellbom P
(2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Molecular Biology 59, 469–484.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Alleva K,
Niemietz CM,
Maurel C,
Parisi M,
Tyerman SD, Amodeo G
(2006) Plasma membrane of Beta vulgaris storage root shows high water channel activity regulated by cytoplasmic pH and a dual range of calcium concentrations. Journal of Experimental Botany 57, 609–621.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Altschul SF,
Madden TL,
Schaffer AA,
Zhang JH,
Zhang Z,
Miller W, Lipman DJ
(1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Baiges I,
Schaffner AR, Mas A
(2001) Eight cDNA encoding putative aquaporins in Vitis hybrid Richter-110 and their differential expression. Journal of Experimental Botany 52, 1949–1951.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Barkla BJ,
Vera-Estrella R,
Pantoja O,
Kirch HH, Bohnert HJ
(1999) Aquaporin localization – how valid are the TIP and PIP labels? Trends in Plant Science 4, 86–88.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Biela A,
Grote K,
Otto B,
Hoth S,
Hedrich R, Kaldenhoff R
(1999) The Nicotiana tabacum plasma membrane aquaporin NtAQP1 is mercury-insensitive and permeable for glycerol. The Plant Journal 18, 565–570.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Borstlap AC
(2002) Early diversification of plant aquaporins. Trends in Plant Science 7, 529–530.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Chaumont F,
Barrieu F,
Jung R, Chrispeels MJ
(2000) Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiology 122, 1025–1034.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Chaumont F,
Barrieu F,
Wojcik E,
Chrispeels MJ, Jung R
(2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiology 125, 1206–1215.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Danielson JAH, Johanson U
(2008) Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biology 8, 45.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Endler A,
Reiland S,
Gerrits B,
Schmidt UG,
Baginsky S, Martinoia E
(2009)
In vivo phosphorylation sites of barley tonoplast proteins identified by a phosphoproteomic approach. Proteomics 9, 310–321.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Engel A,
Fijiyoshi Y, Agre P
(2000) The importance of aquaporin water channel protein structures. EMBO Journal 19, 800–806.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Fetter K,
Van Wilder V,
Moshelion M, Chaumont F
(2004) Interactions between plasma membrane aquaporins modulate their water channel activity. The Plant Cell 16, 215–228.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Fischer M, Kaldenhoff R
(2008) On the pH regulation of plant aquaporins. The Journal of Biological Chemistry 283, 33 889–33 892.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Forrest KL, Bhave M
(2008) The PIP and TIP aquaporins in wheat form a large and diverse family with unique gene structures and functionally important features. Functional & Integrative Genomics 8, 115–133.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Fouquet R,
Leon C,
Ollat N, Barrieu F
(2008) Identification of grapevine aquaporins and expression analysis in developing berries. Plant Cell Reports 27, 1541–1550.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Franks TK,
Powell KS,
Choimes S,
Marsh E,
Iocco P,
Sinclair BJ,
Ford CM, van Heeswijck R
(2006) Consequences of transferring three sorghum genes for secondary metabolite (cyanogenic glucoside) biosynthesis to grapevine hairy roots. Transgenic Research 15, 181–195.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Fu DX,
Libson A,
Miercke LJW,
Weitzman C,
Nollert P,
Krucinski J, Stroud RM
(2000) Structure of a glycerol-conducting channel and the basis for its selectivity. Science 290, 481–486.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Gerbeau P,
Guclu J,
Ripoche P, Maurel C
(1999) Aquaporin Nt-TIPa can account for the high permeability of tobacco cell vacuolar membrane to small neutral solutes. The Plant Journal 18, 577–587.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Gonen T,
Sliz P,
Kistler J,
Cheng YF, Walz T
(2004) Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature 429, 193–197.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Gustavsson S,
Lebrun AS,
Norden K,
Chaumont F, Johanson U
(2005) A novel plant major intrinsic protein in Physcomitrella patens most similar to bacterial glycerol channels. Plant Physiology 139, 287–295.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Harries WEC,
Akhavan D,
Miercke LJW,
Khademi S, Stroud RM
(2004) The channel architecture of aquaporin 0 at a 2.2-angstrom resolution. Proceedings of the National Academy of Sciences of the United States of America 101, 14 045–14 050.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Harvengt P,
Vlerick A,
Fuks B,
Wattiez R,
Ruysschaert JM, Homble F
(2000) Lentil seed aquaporins form a hetero-oligomer which is phosphorylated by a Mg2+-dependent and Ca2+-regulated kinase. The Biochemical Journal 352, 183–190.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hedfalk K,
Tornroth-Horsefield S,
Nyblom M,
Johanson U,
Kjellbom P, Neutze R
(2006) Aquaporin gating. Current Opinion in Structural Biology 16, 447–456.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hill WG,
Southern NM,
MacIver B,
Potter E,
Apodaca G,
Smith CP, Zeidel ML
(2005) Isolation and characterization of the Xenopus oocyte plasma membrane: a new method for studying activity of water and solute transporters. American Journal of Physiology. Renal Physiology 289, F217–F224.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hunt AG
(1994) Messenger-RNA 3′ end formation in plants. Annual Review of Plant Physiology and Plant Molecular Biology 45, 47–60.
|
CAS |

Jahn TP,
Moller ALB,
Zeuthen T,
Holm LM,
Klaerke DA,
Mohsin B,
Kuhlbrandt W, Schjoerring JK
(2004) Aquaporin homologues in plants and mammals transport ammonia. FEBS Letters 574, 31–36.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Jaillon O,
Aury JM,
Noel B,
Policriti A, Clepet C ,
et al
.
(2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Johanson U,
Karlsson M,
Johansson I,
Gustavsson S,
Sjovall S,
Fraysse L,
Weig AR, Kjellbom P
(2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiology 126, 1358–1369.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Johansson I,
Larsson C,
Ek B, Kjellbom P
(1996) The major integral proteins of spinach leaf plasma membranes are putative aquaporins and are phosphorylated in response to Ca2+ and apoplastic water potential. The Plant Cell 8, 1181–1191.
|
CAS |
Crossref |
PubMed |

Johansson I,
Karlsson M,
Shukla VK,
Chrispeels MJ,
Larsson C, Kjellbom P
(1998) Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. The Plant Cell 10, 451–459.
|
CAS |
Crossref |
PubMed |

Jones DT,
Taylor WR, Thornton JM
(1992) The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences 8, 275–282.
|
CAS |
PubMed |

Kamiya T,
Tanaka M,
Mitani N,
Ma JF,
Maeshima M, Fujiwara T
(2009) NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. Journal of Biological Chemistry 284, 2114–2120.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Kammerloher W,
Fischer U,
Piechottka GP, Schaffner AR
(1994) Water channels in the plant plasma-membrane cloned by immunoselection from a mammalian expression system. The Plant Journal 6, 187–199.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Krieg PA, Melton DA
(1984) Functional messenger-RNAs are produced by SP6 invitro transcription of cloned cDNAs. Nucleic Acids Research 12, 7057–7070.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Liman ER,
Tytgat J, Hess P
(1992) Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron 9, 861–871.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Loque D,
Ludewig U,
Yuan LX, von Wiren N
(2005) Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiology 137, 671–680.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ma JF,
Tamai K,
Yamaji N,
Mitani N,
Konishi S,
Katsuhara M,
Ishiguro M,
Murata Y, Yano M
(2006) A silicon transporter in rice. Nature 440, 688–691.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Mahdieh M,
Mostajeran A,
Horie T, Katsuhara M
(2008) Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum plants. Plant & Cell Physiology 49, 801–813.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Marin-Olivier M,
Chevalier T,
Fobis-Loisy I,
Dumas C, Gaude T
(2000) Aquaporin PIP genes are not expressed in the stigma papillae in Brassica oleracea. The Plant Journal 24, 231–240.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Maurel C,
Verdoucq L,
Luu DT, Santoni V
(2008) Plant aquaporins: membrane channels with multiple integrated functions. Annual Review of Plant Biology 59, 595–624.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Mitsuoka K,
Murata K,
Walz T,
Hirai T,
Agre P,
Heymann JB,
Engel A, Fujiyoshi Y
(1999) The structure of aquaporin-1 at 4.5-angstrom resolution reveals short alpha-helices in the center of the monomer. Journal of Structural Biology 128, 34–43.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Moshelion M,
Becker D,
Biela A,
Uehlein N,
Hedrich R,
Otto B,
Levi H,
Moran N, Kaldenhoff R
(2002) Plasma membrane aquaporins in the motor cells of Samanea saman: diurnal and circadian regulation. The Plant Cell 14, 727–739.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Murata K,
Mitsuoka K,
Hirai T,
Walz T,
Agre P,
Heymann JB,
Engel A, Fujiyoshi Y
(2000) Structural determinants of water permeation through aquaporin-1. Nature 407, 599–605.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Neely JD,
Christensen BM,
Nielsen S, Agre P
(1999) Heterotetrameric composition of aquaporin-4 water channels. Biochemistry 38, 11 156–11 163.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Nemeth-Cahalan KL, Hall JE
(2000) pH and calcium regulate the water permeability of aquaporin 0. Journal of Biological Chemistry 275, 6777–6782.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Nemeth-Cahalan KL,
Kalman K, Hall JE
(2004) Molecular basis of pH and Ca2+ regulation of aquaporin water permeability. Journal of General Physiology 123, 573–580.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Picaud S,
Becq F,
Dedaldechamp F,
Ageorges A, Delrot S
(2003) Cloning and expression of two plasma membrane aquaporins expressed during the ripening of grape berry. Functional Plant Biology 30, 621–630.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Prak S,
Hem S,
Boudet J,
Viennois G,
Sommerer N,
Rossignol M,
Maurel C, Santoni V
(2008) Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins. Molecular & Cellular Proteomics 7, 1019–1030.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Preston GM,
Carroll TP,
Guggino WB, Agre P
(1992) Appearance of water channels in Xenopus oocytes expressing red-cell Chip28 protein. Science 256, 385–387.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Quigley F,
Rosenberg JM,
Shachar-Hill Y, Bohnert HJ
(2002) From genome to function: the Arabidopsis aquaporins. Genome Biology 3, research0001.1–research0001.17.
| Crossref | GoogleScholarGoogle Scholar |

Sade N,
Vinocur BJ,
Diber A,
Shatil A,
Ronen G,
Nissan H,
Wallach R,
Karchi H, Moshelion M
(2009) Improving plant stress tolerance and yield production: is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytologist 181, 651–661.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Saitou N, Nei M
(1987) The neighbor-joining method – a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425.
|
CAS |
PubMed |

Sakr S,
Alves G,
Morillon RL,
Maurel K,
Decourteix M,
Guilliot A,
Fleurat-Lessard P,
Julien JL, Chrispeels MJ
(2003) Plasma membrane aquaporins are involved in winter embolism recovery in walnut tree. Plant Physiology 133, 630–641.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Sakurai J,
Ishikawa F,
Yamaguchi T,
Uemura M, Maeshima M
(2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant & Cell Physiology 46, 1568–1577.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Santoni V,
Gerbeau P,
Javot H, Maurel C
(2000) The high diversity of aquaporins reveals novel facets of plant membrane functions. Current Opinion in Plant Biology 3, 476–481.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Savage DF,
Egea PF,
Robles-Colmenares Y,
O’Connell JD, Stroud RM
(2003) Architecture and selectivity in aquaporins: 2.5 Å X-ray structure of aquaporin Z. PLoS Biology 1, 334–340.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Schwede T,
Kopp J,
Guex N, Peitsch MC
(2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Research 31, 3381–3385.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Secchi F,
Lovisolo C,
Uehlein N,
Kaldenhoff R, Schubert A
(2007) Isolation and functional characterization of three aquaporins from olive (Olea europaea L.). Planta 225, 381–392.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Shi LB, Verkman AS
(1996) Selected cysteine point mutations confer mercurial sensitivity to the mercurial-insensitive water channel MIWC/AQP-4. Biochemistry 35, 538–544.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Takano J,
Wada M,
Ludewig U,
Schaaf G,
von Wiren N, Fujiwara T
(2006) The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. The Plant Cell 18, 1498–1509.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tamura K,
Dudley J,
Nei M, Kumar S
(2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Tanaka M,
Wallace IS,
Takano J,
Roberts DM, Fujiwara T
(2008) NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. The Plant Cell 20, 2860–2875.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Törnroth-Horsefield S,
Wang Y,
Hedfalk K,
Johanson U,
Karlsson M,
Tajkhorshid E,
Neutze R, Kjellbom P
(2006) Structural mechanism of plant aquaporin gating. Nature 439, 688–694.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tournaire-Roux C,
Sutka M,
Javot H,
Gout E,
Gerbeau P,
Luu DT,
Bligny R, Maurel C
(2003) Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425, 393–397.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Troggio M,
Vezzulli S,
Pindo M,
Malacarne G,
Fontana P,
Moreira FM,
Costantini L,
Grando MS,
Viola R, Velasco R
(2008) Beyond the genome, opportunities for a modern viticulture: a research overview. American Journal of Enology and Viticulture 59, 117–127.
|
CAS |

Tyerman SD,
Niemietz CM, Bramley H
(2002) Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant, Cell & Environment 25, 173–194.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Uehlein N,
Lovisolo C,
Siefritz F, Kaldenhoff R
(2003) The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425, 734–737.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Vandeleur RK,
Mayo G,
Shelden MC,
Gilliham M,
Kaiser BN, Tyerman SD
(2009) The role of plasma membrane intrinsic protein aquaporins in water transport through roots: diurnal and drought stress responses reveal different strategies between isohydric and anisohydric cultivars of grapevine. Plant Physiology 149, 445–460.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Velasco R,
Zharkikh A,
Troggio M,
Cartwright DA, Cestaro A ,
et al
.
(2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2, e1326.
| Crossref |
PubMed |

Wallace IS,
Choi WG, Roberts DM
(2006) The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochimica et Biophysica Acta (BBA) – Biomembranes 1758, 1165–1175.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Weig AR, Jakob C
(2000) Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NLM2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Letters 481, 293–298.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Whiteman SA,
Nuhse TS,
Ashford DA,
Sanders D, Maathuis FJM
(2008) A proteomic and phosphoproteomic analysis of Oryza sativa plasma membrane and vacuolar membrane. The Plant Journal 56, 146–156.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Zardoya R, Villalba S
(2001) A phylogenetic framework for the aquaporin family in eukaryotes. Journal of Molecular Evolution 52, 391–404.
|
CAS |
PubMed |

Zelazny E,
Borst JW,
Muylaert M,
Batoko H,
Hemminga MA, Chaumont F
(2007) FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proceedings of the National Academy of Sciences of the United States of America 104, 12 359–12 364.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Zhou Y,
Setz N,
Niemietz C,
Qu H,
Offler CE,
Tyerman SD, Patrick JW
(2007) Aquaporins and unloading of phloem-imported water in coats of developing bean seeds. Plant, Cell & Environment 30, 1566–1577.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |




