The convergent evolution of aluminium resistance in plants exploits a convenient currency
Peter R. Ryan A B and Emmanuel Delhaize AA CSIRO Plant Industry, GPO Box 1600, Canberra, ACT 2601, Australia.
B Corresponding author. Email: peter.ryan@csiro.au
This paper is part of an ongoing series: ‘The Evolution of Plant Functions’.
Functional Plant Biology 37(4) 275-284 https://doi.org/10.1071/FP09261
Submitted: 30 October 2009 Accepted: 22 January 2010 Published: 26 March 2010
Abstract
Suspicions that soluble aluminium (Al) is detrimental to plant growth were reported more than 100 years ago. The rhizotoxicity of Al3+ is now accepted as the major limitation to plant production on acidic soils. Plants differ in their susceptibility to Al3+ toxicity and significant variation can occur within species, even in some major crops. The physiology of Al3+ resistance in some species has been understood for 15 years but the molecular biology has been elucidated only recently. The first gene controlling Al3+ resistance was cloned from wheat (Triticum aestivum L.) in 2004 but others have now been identified in Arabidopsis, barley (Hordeum vulgare L.), rye (Secale cereale L.), sorghum (Sorghum bicolour (L.) Moench) and rice (Oryza sativa L.) with strong additional candidates in wheat and oilseed rape (Brassica napus L.). These genes confer resistance in different ways, but one mechanism occurs in nearly all species examined so far. This mechanism relies on the release of organic anions from roots which bind with the harmful Al3+ cations in the apoplast and detoxify them. The genes controlling this response come from at least two distinct families, suggesting that convergent evolution has occurred. We discuss the processes driving this convergence of protein function and offer opinions for why organic anions are central to the mechanisms of resistance in disparate species. We propose that mutations which modify protein expression or their activation by Al3+ have played important roles in co-opting different transport proteins from other functions.
Additional keywords: acid soil, aluminum, anion channel, citrate, malate, tolerance, toxicity.
Al3+ resistance and tolerance in plants
Most polyvalent anions and cations are harmful to plants at low micromolar concentrations (Kinraide 1991, 1994). They are capable of rapidly inhibiting root growth and damaging cells at the root apex but the mechanisms of their toxicity are not fully understood. Trivalent cations, including Al3+, affect cellular functions through an array of intracellular and extracellular interactions which include blocking ion channels, reducing Ca2+ and Mg2+ uptake, competing with Ca2+ for essential binding sites in the apoplast, altering cytoskeletal structure, binding with DNA, disrupting signal transduction pathways and triggering oxidative stress responses (Taylor 1988a; Matsumoto 2000; Yamamoto et al. 2003; Kochian et al. 2004).
Plants encounter Al3+ at harmful concentrations more frequently than any other polyvalent cation. Aluminium is the third most common element in the earth’s crust and acidic conditions accelerate its release from soil minerals into the soil solution. Since 30% of arable lands have a pH of less than 5.5, Al3+ toxicity is an important limitation to plant production and a prevailing pressure for plant adaptation. Some plant species have evolved mechanisms that allow them to survive acid soils better than others. Indeed, genotypes within species can even differ in their ability to withstand Al3+ (Foy 1988). Breeders have exploited this variation to develop cultivars better adapted to acid soils (Garvin and Carver 2003).
The mechanisms that plants have evolved to cope with Al3+ stress can be broadly divided into two main strategies: tolerance mechanisms and resistance or exclusion mechanisms although the divisions between these can be blurry (Taylor 1991; Kochian et al. 2004; Hiradate et al. 2007). Tolerance mechanisms enable plants to safely accommodate Al3+ once it enters the symplast either by chelating it in the cytosol to form harmless complexes or by sequestering it to organelles where it cannot disrupt metabolism. Tolerance mechanisms appear to be common in species endemic to regions with acid soils (e.g. the tropics) where the ability to cope with Al3+ stress is a prerequisite for survival. Examples include tea (Camellia sinensis), buckwheat (Fagopyrum esculentum), Melostoma, and Hydrangea sp., all of which accumulate high concentrations of Al in their leaves (Ma et al. 2001).
Resistance or exclusion mechanisms prevent Al3+ from accumulating in the symplast and minimise harmful interactions with the plasma membrane, cell wall or other targets in the apoplast. These mechanisms rely on root exudates to bind and detoxify the cations in the apoplast (Delhaize et al. 1993), on transport systems to export Al from the symplast or on the capacity to repair damage caused by the Al in the cell wall (Taylor 1991; Huang et al. 2009). Our understanding of resistance mechanisms has progressed more rapidly than tolerance mechanisms because they operate in many common crops (wheat, sorghum, maize, soybean, barley) as well as the model species Arabidopsis and rice and because one or two genes explain most of the phenotypic variation within some of these species. Certainly in wheat, a single locus explains most of the variation in resistance, which led to the conclusion that the trait was not an original condition of this species but appeared more recently in its evolution (Garvin and Carver 2003). Indeed, resistance in wheat appears to be more the exception than the rule. No substantial resistance has been detected in the tetraploid progenitor of hexaploid wheat, Triticum turgidum (Slootmaker 1974; Berzonsky and Kimber 1986; Cosic et al. 1994), and a moderate level of resistance has only recently been identified in the diploid progenitor of hexaploid wheat, Aegilops tauschii (P. R. Ryan and E. Delhaize, unpubl. data).
In summary, Al3+ toxicity is a major selection pressure for plant evolution and many species have evolved tolerance and resistance mechanisms to improve their survival. Some plants appear to display one type of mechanism only (wheat and barley) but others display resistance and tolerance mechanisms which may be additive. Arabidopsis appears to have at least two distinct mechanisms relying, first, on the release of different organic anions from roots that bind with Al3+ (resistance; Hoekenga et al. 2006; Liu et al. 2009) and, second, on the redistribution of Al3+ in the plant (tolerance; Larsen et al. 2005, 2007). Similarly, Fagopyrum esculentum (buckwheat) releases organic anions from its roots (resistance; Zheng et al. 1998) and safely accumulates high concentrations in its leaves (tolerance; Ma et al. 1998).
Al3+ resistance in diverse species relies on the efflux of organic anions
This review will focus on a mechanism of Al3+ resistance widely spread in the plant kingdom which is associated with the efflux of organic anions from roots. The species using this mechanism represent a range of families including the Poaceae (e.g. wheat, barley, sorghum, maize, rye), Araceae (e.g. taro), Polygonaceae (e.g. buckwheat), Brassicaceae (e.g. Arabidopsis) and the Fabaceae (e.g. soybean). Anion efflux is generally restricted to the root apices, the regions most susceptible to Al3+ toxicity (Ryan et al. 1993; Sivaguru and Horst 1998) and the anions released vary from species to species. Malate and citrate are most common, but oxalate efflux has also been detected in a few species (Ma et al. 2001; Ryan et al. 2001; Kochian et al. 2004). All three anions form complexes with Al3+ that are less harmful than the free Al3+ cations and not readily adsorbed by roots. Importantly, the organic anions are not released continuously from the roots in most cases but require Al3+ to trigger the response.
A case of convergent evolution
The first gene controlling Al3+ resistance in plants was isolated from wheat six years ago (Sasaki et al. 2004). The TaALMT1 (Triticum aestivum aluminium-activated malate transporter) gene encodes a member of the ALMT family that consists of membrane-bound proteins (Delhaize et al. 2007). TaALMT1 is located in the plasma membrane and functions as an Al3+-activated anion channel, releasing malate from root cells (Yamaguchi et al. 2005; Zhang et al. 2008). This loss of malate will not necessarily deplete the concentration in the root cells because malate released can be replaced by the continual synthesis of new acids. This can even occur in excised tissue. For example, in one study the cumulative loss of malate over a 4-h period from excised root apices was 3-fold greater than the initial malate content of the tissue (Ryan et al. 1995).
Soon after this gene was described several other members of the ALMT family were shown to contribute to the Al3+ resistance of cereal and non-cereal species in a similar manner (Table 1). These discoveries were exciting at the time because it appeared as though a single gene family controlled Al3+ resistance in a diverse range of species (Magalhaes 2006). However, the model soon required revision after the major resistance genes in sorghum and barley were mapped and sequenced. Aluminium resistance in these species relies on citrate efflux and the proteins involved were not ALMTs but members of a completely different family of proteins named the multi-drug and toxic compound extrusion (MATE) family (Furukawa et al. 2007; Magalhaes et al. 2007; Wang et al. 2007). Other MATE genes were subsequently shown to contribute to the Al3+ resistance of Arabidopsis (Liu et al. 2009) and, probably, wheat (Ryan et al. 2009).
Therefore, at least two gene families separately control the release of malate and citrate, sometimes in the same species. The Arabidopsis genome alone contains ~14 ALMT genes (Delhaize et al. 2007) and 58 MATE genes (Hvorup et al. 2003). The efflux of oxalate from other species may be controlled by a third, as yet, unidentified family. The finding that different gene families confer Al3+ resistance via the same general mechanism is indicative of convergent evolution as suggested previously (Delhaize et al. 2007; Liu et al. 2009). In the remainder of this review we discuss why the efflux of organic anions has emerged as a major mechanism for Al3+ resistance in different gene families.
Why are malate and citrate the common currencies of Al3+ resistance?
Malate and citrate are cheap to synthesise
The stability of the [Al3+:malate] and [Al3+:citrate] complexes is central to their role in protecting plants from Al3+ toxicity. However, inorganic compounds like phosphate and other organic compounds such as cyclic hydroxamates can also form stable complexes with Al3+ (Taylor 1988b; Matsumoto 2000; Kochian et al. 2004; Poschenrieder et al. 2005). Indeed the acidic peptides poly-L-glutamate and poly-L-aspartate are able to protect pollen tube growth and enzyme reactions from Al3+ even more effectively than citrate in some conditions (Konishi et al. 1988; Putterill and Gardner 1988). The same is likely to be true for other proteins and secondary metabolites that possess carboxyl residues or multiple aldehyde and ketone groups capable of chelating Al3+.
Why then have organic anions emerged as the favoured currency for independently evolved mechanisms of Al3+ resistance? The answer may lie in the economy of these small organic compounds. Malate and citrate are ubiquitous in living cells and metabolically cheap to synthesise. These organic anions can almost be considered the ‘small change’ of cellular metabolism. The synthesis of other types of compounds such as some secondary metabolites, polypeptides or even single amino acids requires minerals from the soil and 4–6 carbon compounds as precursors to initiate much longer pathways. Those compounds are necessarily more metabolically ‘expensive’ and demanding of cellular resources. The synthesis of glutamate, for example, requires the uptake of nitrogen from the soil, its reduction to ammonium, a pool of α-ketoglutarate precursors from the tricarboxylic acid cycle and NADPH to drive synthesis.
Malate and citrate are ubiquitous in living cells
Malate is among the most prevalent anions in higher plants with vacuolar concentrations exceeding 200 mM in species that undergo crassulacean acid metabolism (Luttge 1987). The sometimes large and dynamic pools of malate and citrate in most cells reflect their central role in the metabolism of all living organisms (Lance and Rustin 1984). They participate in key anabolic and catabolic pathways including the tricarboxylic acid cycle, the glyoxylate cycle, C4 photosynthetic carbon reduction and crassulacean acid metabolism. Furthermore they contribute directly to nutrient acquisition (phosphorus and iron, Ryan et al. 2001) and osmotic adjustment (guard cells, Fernie and Martinoia 2009) and help to maintain electroneutrality during periods of cation absorption (Osmond 1976; Ryan et al. 2001). Malate is also involved in regulating cytosolic pH by shuttling across the tonoplast (Kovermann et al. 2007) and by participating in the biochemical ‘pH stat’ reactions (Sakano 1998). The pH-stat model proposes that a series of enzymes finely regulate pH through reactions which produce or consume protons. The combined activities of phosphoenolpyruvate carboxylase (pH optimum ~7.8), malate dehydrogenase and malic enzyme (pH optimum <7.0) buffer cytosolic pH by balancing the synthesis and degradation of malate as well as influencing the rate of glycolysis. These reactions bypass the standard pathway supplying pyruvate for the TCA cycle by allowing oxidation of four-carbon acids in the absence of pyruvate. This shortcut is also useful during periods of phosphate deficiency because the phosphoenolpyruvate carboxylase reaction releases inorganic phosphate (Theodorou and Plaxton 1993).
Malate and citrate need to be shuffled between subcellular compartments
To satisfy their diverse roles, efficient transport systems are required to shuttle malate and citrate in and out of cells and between the subcellular compartments. The movement of these substrates across membranes needs to be mediated by specific membrane-bound proteins. Some of these proteins catalyse energetically passive reactions, some expend energy directly or indirectly and move ions against electrochemical gradients, and others facilitate the coordinated symport or antiport with other substrates. The genes encoding some of these transport proteins have been cloned (Reumann and Weber 2006; Martinoia et al. 2007) but many more are yet to be identified. Indeed the 20 or so metabolite transporters now identified and characterised in plastids represent less than 20% of the total predicted to exist from physiological and bioinformatic analyses (Weber and Fischer 2007). The finding that the AtALMT9 protein in Arabidopsis resides on the tonoplast demonstrates that some members of the ALMT family function on internal membranes as well as the plasma membrane (Kovermann et al. 2007).
Hypothesis: the evolution of Al3+ resistance via organic anion efflux arose from mutations that co-opted malate and citrate transport proteins from other functions
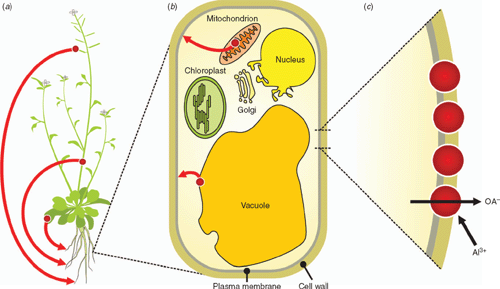
As discussed above, Al3+ resistance is unlikely to have been an early trait for plants that did not evolve on acidic environments. Instead, it would have evolved after their distribution extended into regions with acid soils (Garvin and Carver 2003). We propose that Al3+ resistance arose from mutations that co-opted some of the many malate or citrate transport proteins that originally performed different functions (Fig. 1). For example, if a protein facilitating malate release from a stomatal guard cell in leaves incurs a mutation that redirects or extends its expression to roots, the efflux of malate from the root cells could afford some protection from Al3+ toxicity and provide the plant with a selective advantage on acid soils. This trait could undergo further selection in subsequent generations and become more widely represented in the population if the selection pressure is sustained. A recent study on the Arabidopsis gene AtFRD3 provides ‘in principal’ support for the idea that extending the expression of a gene can confer a new Al3+‐resistance phenotype. AtFRD3 encodes a MATE protein expressed in cells surrounding the root vasculature where it facilitates the efflux of citrate into the xylem to accompany iron movement to the shoots (Rogers and Guerinot 2002; Durrett et al. 2007). Ectopic expression of AtFRD3 with the CaMV35S promoter causes no deleterious effects to the Arabidopsis plants but the resulting constitutive efflux of citrate from the roots enhances Al3+ resistance (Durrett et al. 2007). The original location of AtFRD3 on the plasma membrane of vascular cells makes this protein predisposed to mutations that extend its function to include citrate efflux from roots. Overexpression of HvALMT1 in barley also confers Al3+ resistance through constitutive malate efflux from roots but the transgenic plants show severe stunting and leaf necrosis (Gruber 2009). This deleterious phenotype might be related to the disruption of organic anion homeostasis internally, since the HvALMT1 proteins localise to internal membranes as well as the plasma membrane.
|
Similar outcomes are possible for proteins redirected from other tissues to the roots or from internal membranes to the plasma membrane as long as the protein still functions in its new location. Perhaps the simplest scenario would involve proteins already located on the plasma membrane of root cells which incur mutations that increase their expression or enable their function to be activated by Al3+ (see later). These proteins could be facilitating organic anion release from root cells for other reasons such as osmotic adjustment or nutrient acquisition.
The question of why some species release malate from their roots and others release citrate might not depend on the concentration of organic anions in the tissue or on their biochemistry. Instead, it might simply relate to whether the original function of the transport protein co-opted to perform this new role involved malate transport or citrate transport. This is supported by transgenic experiments that overexpressed the wheat TaALMT1 gene in barley (Delhaize et al. 2004). The relatively small variation in the Al3+ resistance among genotypes of barley relies on Al3+-activated citrate efflux, not malate efflux, yet barley plants expressing TaALMT1 show the customary Al3+-activated malate efflux. This demonstrates that barley is capable of releasing malate from its roots providing a suitable transport mechanism is present.
Presumably it would be necessary for the original function of the protein to be maintained or replaced so that these mutations are not detrimental. If the mutation simply extends the expression of the gene to new tissues or membranes then function could continue in the original location. If the protein is completely relocated then other proteins with similar function (redundancy) need to be able to compensate for their loss. A common source of redundancy in plants is local gene duplication where individual genes within chromosomes are copied. Redundancy can also arise from the hybridisation of entire sets of chromosomes as has occurred in wheat (polyploidisation). Redundancy provides insurance against lethal mutations and enables genes to be changed or re-deployed without adverse effects. Therefore, redundancy can benefit the fitness of organisms by facilitating evolutionary experimentation in changing environments (Otto and Whitton 2000).
Of course, not all transport proteins will be suitable for facilitating the efflux of useful anions from root cells. For instance, the energetics and mechanism of the transport protein, their pH sensitivity or their phosphorylation state are some factors that can affect function. The efflux of malate and citrate from root cells involves the movement of anions from the cytosol to the apoplast. This is an energetically ‘down-hill’ or passive process due to the large negative electrical potential of the cytosol relative to the outside of –100 to –200 mV. Ion channels are one type of transport protein that would be suitable because they facilitate passive ion movement and physiological studies indicate that TaALMT1 functions as an anion channel (Zhang et al. 2008). By contrast, the malate/oxaloacetate antiporter found on the inner mitochondrial membrane is an example of a malate transporter that is unlikely to work on the plasma membrane of root cells unless there is sufficient oxaloacetate in the apoplasm to satisfy the exchange reaction.
What type of mutations could generate these changes in tissue expression or membrane localisation?
Where a protein is finally located in a cell relies on short amino acid motifs called targeting domains. The sequence of these targeting domains and their position on the protein contain information that ensures soluble proteins reach their appropriate subcellular compartment (e.g. plastid, mitochondria, vacuole) and that transport proteins and other membrane-bound enzymes reach their target membranes (e.g. tonoplast, plasma membrane, mitochondrial membrane). Targeting domains occur at the N- and C-terminal ends of the protein and in some cases they are removed once the protein has reached its final destination. Intracellular localisation of a protein can therefore be affected by mutations to the DNA sequences encoding these targeting domains.
By contrast, the level of expression and tissue specificity are controlled by cis regulatory elements generally found in the promoter region of the gene. Cis-regulatory elements are untranscribed DNA sequences that influence when, where, and to what level genes are expressed. Relatively simple mutations in these elements could influence phenotype by redirecting proteins to different tissues or by altering their level of expression. Nucleotide substitutions, deletions or tandem repeats in cis-regulatory elements are common mutations that have the potential to alter gene expression and, consequently, plant phenotype.
Modifications to the target sequences and cis-regulatory elements can influence evolutionary change just as mutations in the coding regions do by changing protein function (Wray et al. 2003). The role of cis mutations and transcriptional regulation in species diversification is receiving increased attention. Its importance has been strengthened by reports directly linking phenotype to gene expression (Wray et al. 2003). We already have evidence that cis mutations have contributed to the evolution of Al3+ resistance in wheat by altering the expression of TaALMT1 and this is discussed in more detail later.
Can this model accommodate other observations on Al3+ resistance?
The cases of STOP1 and ART1
STOP1 in Arabidopsis and ART1 in rice encode C2H2 zinc finger-type transcription factors that control the expression of several genes providing resistance to Al3+ stress (ART1; Yamaji et al. 2009) or to low pH and Al3+ stress (STOP1; Sawaki et al. 2009). STOP1 regulates two genes involved in Al3+-activated malate and citrate efflux (AtALMT1 and AtMATE1) both of which contribute to the Al3+ resistance of Arabidopsis (Table 1; Liu et al. 2009; Sawaki et al. 2009). The involvement of transcription factors such as STOP1 and ART1 can be accommodated by our model. The proposed mutations that divert malate and citrate transport proteins from their original sites of expression or membrane location to the plasma membrane of root cells would not necessarily interfere with any pre-existing regulation by transcription factors. The sequences recognised by transcription factors tend to be short modular regions of the promoter and these will not be affected by changes occurring in the coding region. Those sequences can also be physically separated from mutations to neighbouring regions of the promoter that control expression level or tissue specificity.
Activation and induction by Al3+
The ALMT and MATE genes conferring Al3+ resistance are constitutively expressed in some plants and induced by Al3+ treatment in others. However, regardless of whether or not the genes are induced by Al3+, nearly all the proteins they encode require external Al3+ to activate their function and release organic anions (Table 1). It is unclear how this activation occurs or even which form of Al is responsible for activation because soluble Al exists in a pH-dependent equilibrium among several ionic species (Al3+, Al(OH)2+, Al(OH)2+ and Al(OH)4–; Kinraide 1991). Nevertheless Al3+, the most likely candidate (Ryan et al. 1995), could directly trigger efflux, by interacting with the proteins, or indirectly, by first initiating a signal cascade or stress response, which then activates transport activity (Ryan et al. 2001). The direct-trigger explanation is more likely given this same dependence for Al3+ is observed in every heterologous expression system used to examine the function of these proteins. For example, TaALMT1 has now been expressed in a range of plants (e.g. rice, barley, tobacco, Arabidopsis), cell cultures (tobacco suspension cells) and animal cells (Xenopus laevis oocytes) using constitutive promoters or cRNA injection and, in every case, addition of Al3+ to the bathing solution increases malate efflux. It seems simpler for Al3+ to directly activate anion efflux than to expect that all those different cell types share pathways capable of first interacting with Al3+ and then transducing that signal to activate function. An exception to this dependency on Al3+ is in wheat where a constitutive efflux of citrate contributes to the Al3+ resistance in a few genotypes (Table 1; Ryan et al. 2009).
We view the induction of protein expression and the activation of protein function by Al3+ as useful traits for minimising unnecessary carbon loss in the absence of toxicity. Since Al3+ poses little problem in soils with pH >5.5 it would be wasteful for plants to continually release organic anions, even at a small metabolic cost, when there is no benefit. This requirement for Al3+ to activate efflux would also help stabilise the trait in a population because the resistance gene would not be selected against in non-acid conditions when there is no penalty to fitness.
In some species Al3+ both induces expression of the ALMT and MATE genes and activates protein function (Table 1). Why should both responses occur in a single species when these dual ‘safety’ mechanisms appear redundant? Part of the explanation might be that the induction of expression is a general response to stress that predates the involvement of the transport protein in Al3+ resistance whereas the capacity for the protein to be activated by Al3+ is a trait acquired more recently. In other words, the expression of these genes might have already responded to some other stress before they were co-opted from their original function to contribute to Al3+ resistance. There is some evidence for this because the major Al3+ resistance genes in Arabidopsis, AtALMT1 and AtMATE1, are induced by other stimuli. Both genes are partly induced by low pH (Kobayashi et al. 2007; Liu et al. 2009) and AtALMT1 is strongly induced by foliar infection with Pseudomonas syringae (Rudrappa et al. 2008). Similarly, the expression of BnALMT1-1 and BnALMT1-2 in oilseed rape is induced by metal ions other than Al3+ (Ligaba et al. 2006). Therefore, in some cases at least, the Al3+-dependent increase in expression might reflect the induction by general stress and not a specific response to Al3+ toxicity. If so, the activation by Al3+ becomes an even more important point of control because, regardless of the stress inducing ALMT or MATE expression, anion efflux will only occur when toxic Al3+ is present in the soil.
If the co-opted ALMT or MATE proteins were originally located on intracellular membranes or in shoot tissues, then the requirement for Al3+ to activate their function is more likely to have appeared after the proteins were relocated to the plasma membrane of root cells. The reason for this is that intracellular proteins and leaf tissues are not usually exposed to soluble Al3+. Even if Al3+ manages to enter the symplast, the alkaline pH and plethora of potential binding compounds maintain the concentration of free Al3+ cations extremely low. It is doubtful that proteins are specifically activated by a ligand they rarely, if ever, encounter. If we conclude then, that activation by Al3+ evolved after the proteins were co-opted to their new role then we are also forced to conclude that this change occurred independently in the ALMT and MATE families. The surprising corollary to this position is that relatively simple changes in the amino acid sequence of very different proteins can alter their function in a similar manner (i.e. allow them to be activated by Al3+). If so, we may get some idea what these changes are by comparing the sequence of proteins that do or do not show activation in order to identify residues associated with this phenotype. Mutational analysis could then confirm the importance of any candidate residues. Nevertheless it remains possible that activation of the proteins by Al3+ did not evolve as described above but is a non-specific response. For instance, the highly charged Al3+ ions might simply induce conformational changes in these proteins that alter their activity. Previous reports showing that other trivalent cations, such as erbium, can also activate TaALMT1, albeit to lesser extent, support this idea by demonstrating that activation is not absolutely specific for Al3+ (Kataoka et al. 2002; Delhaize et al. 2004). One last example shows that it is even possible to totally circumvent this requirement for Al3+. The foliar infection of Arabidopsis plants with a strain of Pseudomonas syringae not only induces AtALMT1 expression but triggers malate efflux (Rudrappa et al. 2008). This intriguing response, which reportedly encourages the proliferation of beneficial microorganisms to the rhizosphere, suggests that AtALMT1 may have multiple functions. It also begs the question of whether AtALMT1 was originally co-opted from a plant-microbe function to contribute to Al3+ resistance or vise versa.
Supportive evidence
Hypotheses which attempt to explain evolutionary processes are inherently difficult to prove or disprove. Nevertheless, we can ask what type of data would be consistent with the hypothesis and see if those data are available or whether they can be obtained. For instance, what evidence would support the idea proposed here that mutations in cis regulatory elements may have altered the level of protein expression or its tissue specificity? The demonstration that differences in promoter sequence can change the pattern or level of gene expression would support the hypothesis.
One example is illustrated by the perfect tandem repeats that occur in the promoter of TaALMT1 in most Al3+-resistant wheats (Sasaki et al. 2006). These repeats occur as duplications or triplications 31–803 bp long that are associated with increased expression of TaALMT1 and greater malate efflux (Raman et al. 2008). Four of the tandem repeat patterns appear to have evolved independently of one another (Delhaize et al. 2007) with at least two conferring enhanced TaALMT1 expression as demonstrated experimentally by transforming rice with several promoter regions fused to the reporter gene encoding green fluorescent protein (E. Delhaize and P. R. Ryan, unpubl. data). These tandem repeats appear to have arisen after the appearance of T. aestivum some 10 000 years ago (Dubcovsky and Dvorak 2007) because they have not been detected in Ae. tauschii the D-genome progenitor of hexaploid wheat. Polyploidy may have been helpful in fixing these mutations in subpopulations of T. aestivum as discussed above. How these tandem repeats formed is not known but may be due to the activation of mobile elements associated with allopolyploidy in cereals (Kashkush et al. 2003). For instance, transposable elements such as Helitrons possess the rolling-circle machinery capable of generating these types of tandem repeats as suggested by Piffanelli et al. (2004). Illegitimate recombination is another mechanism that can generate duplications but the signature short direct repeats that usually flank these duplications (Wicker et al. 2007) are absent from the tandem repeats in the TaALMT1 promoter. Once a region of the genome is duplicated by whatever means, additional copies can be generated by unequal crossing over.
Variations in the promoter regions of MATE genes are also associated with enhanced Al3+ resistance. Major genes for Al3+ resistance in sorghum (SbMATE) and barley (HvMATE, also named HvAACT1) encode MATE proteins that are activated by Al3+ and efflux citrate (Furukawa et al. 2007; Magalhaes et al. 2007; Wang et al. 2007). The sorghum SbMATE promoter possesses multiple insertions of a tourist-like miniature inverted repeat transposable element (MITE) and the number of MITES correlates with the level of expression and Al3+ resistance (Magalhaes et al. 2007). However, direct evidence, such as those obtained from reporter-gene studies, showing that these insertions enhance expression level is currently lacking. Similarly, the promoter region of HvMATE in an Al3+-resistant barley cultivar possesses a 1023-bp insertion 4.6 kb upstream of the open reading frame start site that is absent from sensitive cultivars (Fujii et al. 2009). This insertion contains multiple transcription start sites that may be responsible for the high level of HvMATE expression found in Al3+ resistant genotypes.
The above examples described one type of cis mutation that confers a novel phenotype by changing the level of transcription but others can be investigated. The related idea that mutations to the targeting sequences of a protein can generate new phenotypes can be examined by first establishing a correlation between the coding alleles of a gene and Al3+ resistance. We can then test whether allelic variants direct the protein to different cellular membranes when fused to a reporter gene and expressed in a heterologous system. Once again, evidence establishing that certain coding alleles generate new phenotypes via protein targeting would provide additional support to the hypotheses proposed in this review.
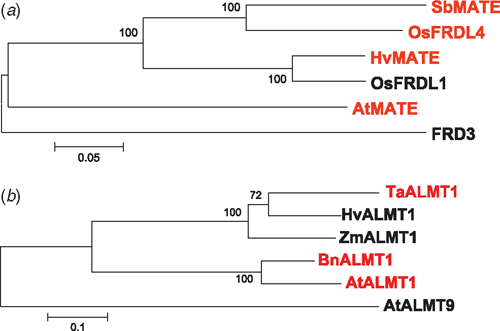
Our model that convergent evolution has resulted from the procurement of different gene families to enhance Al3+ resistance can even be extended to different genes within the same family. For instance, although barley HvMATE, sorghum SbMATE and Arabidposis AtMATE all belong to the same gene family, they appear not to have originated from a common ancestral MATE gene since they do not cluster as a group distinct from other MATEs that confer constitutive citrate efflux (Fig. 2a). The same conclusion can be drawn from the ALMTs because among the proteins characterised so far those involved in Al3+ resistance do not cluster together in a phylogenetic tree (Fig. 2b). We know that ALMT proteins occur on different membranes and have functional roles beyond Al3+ resistance. Both ZmALMT1 and HvALMT1 have greater similarity to TaALMT1 than AtALMT1 yet neither appears to have a role in Al3+ resistance (Piñeros et al. 2008; Gruber 2009). These observations suggest that two or more genes from each of these families were independently recruited for a role in Al3+ resistance and each independently evolved the capacity for their function to be activated by Al3+. Alternatively, it is also plausible that an ancestral MATE gene did encode a protein that was activated by Al3+ and that OsFRDL4 and AtFRD3 subsequently lost this activation domain to assume roles in the long distant transport of iron (Durrett et al. 2007; Yokosho et al. 2009b). A similar argument for the involvement of ALMT genes in Al3+ resistance has been made previously (Delhaize et al. 2007) despite apparent conservation of Al3+ resistance genes between monocots and dicots (Magalhaes 2006). The rye genes (ScALMT1-M39.1 and M39.2) which are closely related to TaALMT1 and appear to confer resistance based on Al3+-activated efflux of organic anions (Table 1) probably share a common origin. However, the more distantly-related Arabidopsis gene AtALMT1, which is not the most similar member of this family in that genome to TaALMT1, is likely to have developed a role in Al3+ resistance independently (Delhaize et al. 2007).
|
Summary
The emergence of Al3+ resistance in plants appears to be a classic example of convergent evolution. This is based on the finding that the same mechanism of Al3+ resistance operating in a range of plants is encoded by distinct gene families. The criteria that make malate and citrate anions the ideal currency of this mechanism include their ability to form stable complexes with Al3+, their prevalence in plant cells, the economy of their synthesis and their requirement to be moved across most cellular membranes by an array of different transport proteins. We propose that the mechanism of Al3+ resistance based on organic anion efflux from root cells evolved relatively recently from mutations that co-opted transport proteins from other locations or other functions. Among the many proteins transporting organic anions across plant-cell membranes, members of the MATE and ALMT families appear most suited and readily recruited to assume roles in Al3+ resistance.
Note added in proof
A relevant paper was published after these proofs were prepared. Maron et al. (2010) identified and characterised two MATE genes that map to major aluminium‐resistance QTLs in maize.
Berzonsky WA, Kimber G
(1986) Tolerance of Triticum species to aluminum. Plant Breeding 97, 275–278.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Collins NC,
Shirley NJ,
Saeed M,
Pallotta M, Gustafson JP
(2008) An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L.). Genetics 179, 669–682.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Cosic T,
Poljak M,
Custic M, Rengel Z
(1994) Aluminum tolerance of durum-wheat germplasm. Euphytica 78, 239–243.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Delhaize E,
Ryan PR, Randall PJ
(1993) Aluminum tolerance in wheat (Triticum aestivum L.) II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiology 103, 695–702.
|
CAS |
PubMed |

Delhaize E,
Ryan PR,
Hebb DM,
Yamamoto Y,
Sasaki T, Matsumoto H
(2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proceedings of the National Academy of Sciences of the United States of America 101, 15249–15254.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Delhaize E,
Gruber BD, Ryan PR
(2007) The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Letters 581, 2255–2262.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Dubcovsky J, Dvorak J
(2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Durrett TP,
Gassmann W, Rogers EE
(2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiology 144, 197–205.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Felsenstein J
(1985) Confidence-limits on phylogenies – an approach using the bootstrap. Evolution 39, 783–791.
| Crossref | GoogleScholarGoogle Scholar |

Fernie AR, Martinoia E
(2009) Malate. Jack of all trades or master of a few? Phytochemistry 70, 828–832.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Foy CD
(1988) Plant adaptation to acid, aluminum-toxic soils. Communications in Soil Science and Plant Analysis 19, 959–987.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Furukawa J,
Yamaji N,
Wang H,
Mitani N,
Murata Y,
Sato K,
Katsuhara M,
Takeda K, Ma JF
(2007) An aluminum-activated citrate transporter in barley. Plant & Cell Physiology 48, 1081–1091.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hiradate S,
Ma JF, Matsumoto H
(2007) Strategies of plants to adapt to mineral stresses in problem soils. Advances in Agronomy 96, 65–132.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Hoekenga OA,
Maron LG,
Piñeros MA,
Cançado GMA, Shaff JE ,
et al
.
(2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 103, 9738–9743.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Huang CF,
Yamaji N,
Mitani N,
Yano M,
Nagamura Y, Ma JF
(2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. The Plant Cell 21, 655–667.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hvorup RN,
Winnen B,
Chang AB,
Jiang Y,
Zhou XF, Saier MH
(2003) The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. European Journal of Biochemistry 270, 799–813.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Kashkush K,
Feldman M, Levy AA
(2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nature Genetics 33, 102–106.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Kataoka T,
Stekelenburg A,
Nakanishi TM,
Delhaize E, Ryan PR
(2002) Several lanthanides activate malate efflux from roots of aluminium-tolerant wheat. Plant, Cell & Environment 25, 453–460.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Kinraide TB
(1991) Identity of the rhizotoxic aluminium species. Plant and Soil 134, 167–178.
|
CAS |

Kinraide TB
(1994) Use of a Gouy–Chapman–Stern model for membrane-surface electrical potential to interpret some features of mineral rhizotoxicity. Plant Physiology 106, 1583–1592.
|
CAS |
PubMed |

Kobayashi Y,
Hoekenga OA,
Itoh H,
Nakashima M,
Saito S,
Shaff JE,
Maron LG,
Piñeros MA,
Kochian LV, Koyama H
(2007) Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiology 145, 843–852.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Kochian LV,
Hoekenga OA, Piñeros MA
(2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorus efficiency. Annual Review of Plant Biology 55, 459–493.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Konishi S,
Ferguson IB, Putterill J
(1988) Effect of acidic polypeptides on aluminium toxicity in tube growth of pollen from tea (Camellia sinensis L.). Plant Science 56, 55–59.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Kovermann P,
Meyer S,
Hortensteiner S,
Picco C,
Scholz-Starke J,
Ravera S,
Lee Y, Martinoia E
(2007) The Arabidopsis vacuolar malate channel is a member of the ALMT family. The Plant Journal 52, 1169–1180.
|
CAS |
PubMed |

Lance C, Rustin P
(1984) The central role of malate in plant-metabolism. Physiologie Vegetale 22, 625–641.
|
CAS |

Larsen PB,
Geisler MJB,
Jones CA,
Williams KM, Cancel JD
(2005) ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. The Plant Journal 41, 353–363.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Larsen PB,
Cancel J,
Rounds M, Ochoa V
(2007) Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225, 1447–1458.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ligaba A,
Katsuhara M,
Ryan PR,
Shibasaka M, Matsumoto H
(2006) The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiology 142, 1294–1303.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Liu JP,
Magalhaes JV,
Shaff J, Kochian LV
(2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. The Plant Journal 57, 389–399.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Luttge U
(1987) Carbon-dioxide and water demand- crassulacean acid metabolism (CAM), a versatile ecological adaptation exemplifying the need for integration in ecophysiological work. New Phytologist 106, 593–629.
| Crossref | GoogleScholarGoogle Scholar |

Ma JF,
Hiradate S, Matsumoto H
(1998) High aluminum resistance in buckwheat. II. Oxalic acid detoxifies aluminum internally. Plant Physiology 117, 753–759.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ma JF,
Ryan PR, Delhaize E
(2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends in Plant Science 6, 273–278.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Magalhaes JV
(2006) Aluminum tolerance genes are conserved between monocots and dicots. Proceedings of the National Academy of Sciences of the United States of America 103, 9749–9750.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Magalhaes JV,
Liu J,
Guimaraes CT,
Lana UGP, Alves VMC ,
et al
.
(2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nature Genetics 39, 1156–1161.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Maron LG,
Pineros MA,
Guimaraes CT,
Magalhaes JV,
Pleiman JK,
Mao C,
Shaff JE,
Belicuas SNJ, Kochian LV
(2010) Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. The Plant Journal 61, 728–740.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Martinoia E,
Maeshima M, Neuhaus HE
(2007) Vacuolar transporters and their essential role in plant metabolism. Journal of Experimental Botany 58, 83–102.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Matsumoto H
(2000) Cell biology of aluminum toxicity and tolerance in higher plants. International Review of Cytology 200, 1–46.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Otto SP, Whitton J
(2000) Polyploid incidence and evolution. Annual Review of Genetics 34, 401–437.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Piffanelli P,
Ramsay L,
Waugh R,
Benabdelmouna A,
D’Hont A,
Hollricher K,
Jorgensen JH,
Schulze-Lefert P, Panstruga R
(2004) A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430, 887–891.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Piñeros MA,
Cançado GMA,
Maron LG,
Lyi SM,
Menossi M, Kochian LV
(2008) Not all ALMT1-type transporters mediate aluminum-activated organic acid responses: the case of ZmALMT1 – an anion-selective transporter. The Plant Journal 53, 352–367.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Poschenrieder C,
Tolrà RP, Barcelo J
(2005) A role for cyclic hydroxamates in aluminium resistance in maize? Journal of Inorganic Biochemistry 99, 1830–1836.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Putterill JJ, Gardner RC
(1988) Proteins with the potential to protect plants from Al3+ toxicity. Biochimica et Biophysica Acta 1988, 137–145.

Raman H,
Ryan PR,
Raman R,
Stodart BJ, Zhang K ,
et al
.
(2008) Analysis of TaALMT1 traces the transmission of aluminum resistance in cultivated common wheat (Triticum aestivum L.). Theoretical and Applied Genetics 116, 343–354.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Reumann S, Weber APM
(2006) Plant peroxisomes respire in the light: some gaps of the photorespiratory C-2 cycle have become filled – others remain. Biochimica et Biophysica Acta – Molecular. Cell Research 1763, 1496–1510.
|
CAS |
Crossref |

Rogers EE, Guerinot ML
(2002) FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. The Plant Cell 14, 1787–1799.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Rudrappa T,
Czymmek KJ,
Pare PW, Bais HP
(2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiology 148, 1547–1556.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ryan PR,
DiTomaso JM, Kochian LV
(1993) Aluminium toxicity in roots: an investigation of spatial sensitivity and the role of the root cap. Journal of Experimental Botany 44, 437–446.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Ryan PR,
Delhaize E, Randall PJ
(1995) Characterisation of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots. Planta 196, 103–110.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Ryan PR,
Delhaize E, Jones DL
(2001) Function and mechanism of organic anion exudation from plant roots. Annual Review of Plant Physiology and Plant Molecular Biology 52, 527–560.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ryan PR,
Raman H,
Gupta S,
Horst WJ, Delhaize E
(2009) A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiology 149, 340–351.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Saitou N, Nei M
(1987) The neighbour-joining method – a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425.
|
CAS |
PubMed |

Sakano K
(1998) Revision of biochemical pH-stat: involvement of alternative pathway metabolisms. Plant & Cell Physiology 39, 467–473.
|
CAS |

Sasaki T,
Yamamoto Y,
Ezaki B,
Katsuhara M,
Ahn SJ,
Ryan PR,
Delhaize E, Matsumoto H
(2004) A wheat gene encoding an aluminum-activated malate transporter. The Plant Journal 37, 645–653.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Sasaki T,
Ryan PR,
Delhaize E,
Hebb DM, Ogihara Y ,
et al
.
(2006) Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant & Cell Physiology 47, 1343–1354.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Sawaki Y,
Iuchi S,
Kobayashi Y,
Kobayashi Y, Ikka T ,
et al
.
(2009) STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiology 150, 281–294.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Sivaguru M, Horst WJ
(1998) The distal part of the transition zone is the most aluminum-sensitive apical root zone of Zea mays L. Plant Physiology 116, 155–163.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Slootmaker LAJ
(1974) Tolerance to high soil acidity in wheat related species, rye and triticale. Euphytica 23, 505–513.
| Crossref | GoogleScholarGoogle Scholar |

Tamura K,
Dudley J,
Nei M, Kumar S
(2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Taylor GJ
(1988b) The physiology of aluminum tolerance in higher plants. Communications in Soil Science and Plant Analysis 19, 1179–1194.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Taylor GJ
(1991) Current views of the aluminum stress response: the physiological basis of tolerance. Current Topics in Plant Biochemistry and Physiology 10, 57–93.
|
CAS |

Theodorou ME, Plaxton WC
(1993) Metabolic adaptations of plant respiration to nutritional phosphate deprivation. Plant Physiology 101, 339–344.
|
CAS |
PubMed |

Wang JP,
Raman H,
Zhou MX,
Ryan PR,
Delhaize E,
Hebb DM,
Coombes N, Mendham N
(2007) High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 115, 265–276.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Weber APM, Fischer K
(2007) Making the connections – the crucial role of metabolite transporters at the interface between chloroplast and cytosol. FEBS Letters 581, 2215–2222.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Wicker T,
Yahiaoui N, Keller B
(2007) Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. The Plant Journal 51, 631–641.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Wray GA,
Hahn MW,
Abouheif E,
Balhoff JP,
Pizer M,
Rockman MV, Romano LA
(2003) The evolution of transcriptional regulation in eukaryotes. Molecular Biology and Evolution 20, 1377–1419.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Yamaguchi M,
Sasaki T,
Sivaguru M,
Yamamoto Y,
Osawa H,
Ahn SJ, Matsumoto H
(2005) Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1). Plant & Cell Physiology 46, 812–816.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Yamaji N,
Huan CF,
Nagao S,
Yano M,
Sato Y,
Nagamura Y, Ma JF
(2009) A zinc finger transcription factor ART1 regulates multiple genes involved in aluminum tolerance in rice. The Plant Cell 21, 3339–3349.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Yamamoto Y,
Kobayashi Y,
Devi SR,
Rikiishi S, Matsumoto H
(2003) Oxidative stress triggered by aluminum in plant roots. Plant and Soil 255, 239–243.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Yokosho K,
Yamaji N,
Ueno D,
Mitani N, Ma JF
(2009b) OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiology 149, 297–305.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Zhang W,
Ryan PR,
Sasaki T,
Yamamoto Y,
Sullivan W, Tyerman SD
(2008) Characterization of the TaALMT1 protein as an Al3+-activated anion channel in transformed tobacco (Nicotiana tabacum L.) cells. Plant & Cell Physiology 49, 1316–1330.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Zheng SJ,
Ma JF, Matsumoto H
(1998) High aluminum resistance in buckwheat. 1. Al-induced specific secretion of oxalic acid from root tips. Plant Physiology 117, 745–751.
| Crossref | GoogleScholarGoogle Scholar | PubMed |




