Investigation of the domestication of common bean (Phaseolus vulgaris) using multilocus sequence data
Sujan Mamidi A D E , Monica Rossi B , Deepti Annam C , Samira Moghaddam A D , Rian Lee A D , Roberto Papa B and Phillip McClean A DA North Dakota State University, Department of Plant Sciences, Fargo, ND 58102, USA.
B Università Politecnica delle Marche, Scienze Ambientali e delle Produzioni Vegetali, Ancona, Italy.
C North Dakota State University, Department of Statistics, Fargo, ND 58102, USA.
D North Dakota State University, Genomics and Bioinformatics Program, Fargo, ND 58102, USA.
E Corresponding author. Emails: sujan_vnv@yahoo.com, sujan.mamidi@gmail.com
Functional Plant Biology 38(12) 953-967 https://doi.org/10.1071/FP11124
Submitted: 21 May 2011 Accepted: 15 September 2011 Published: 7 November 2011
Abstract
Multilocus sequence data collected from domesticated and related wild relatives provides a rich source of information on the effect of human selection on the diversity and adaptability of a species to complex environments. To evaluate the domestication history of common bean (Phaseolus vulgaris L.), multilocus sequence data from landraces representing the various races within the Middle American (MA) and Andean gene pools was evaluated. Across 13 loci, nucleotide diversity was similar between landraces and wild germplasm in both gene pools. The diversity data were evaluated using the approximate Bayesian computation approach to test multiple domestication models and estimate population demographic parameters. A model with a single domestication event coupled with bidirectional migration between wild and domesticated genotypes fitted the data better than models consisting of two or three domestication events in each genepool. The effective bottleneck population size was ~50% of the base population in each genepool. The bottleneck began ~8200 and ~8500 years before present and ended at ~6300 and ~7000 years before present in MA and Andean gene pools respectively. Linkage disequilibrium decayed to a greater extent in the MA genepool. Given the (1) geographical adaptation bottleneck in each wild gene pool, (2) a subsequent domestication bottleneck within each gene pool, (3) differentiation into gene-pool specific races and (4) variable extents of linkage disequilibrium, association mapping experiments for common bean would more appropriately be performed within each genepool.
Additional keywords: ABC approach, association mapping, bottleneck, demography, genepools, linkage disequilibrium, races.
Introduction
Domestication is a complex process in which human usage of plant and animal species has led to morphological and physiological changes that made them genetically different from the wild types and better adapted to different agro-ecosystems (Glémin and Bataillon 2009). Beginning in the Epipalaeolithic and extending into the Neolithic period (13 000–11 000 years ago), cultivation started with just a few plant species as food sources (Fuller 2007). Many morphological and physiological changes were associated with the process of domestication and are termed the ‘domestication syndrome’ (Glémin and Bataillon 2009). The study of domestication as an evolutionary model can identify events associated with the origins of crop species and describe the selective pressures experienced by domesticated taxa.
Common bean (Phaseolus vulgaris L.) is the most important dietary legume in the world because of its high concentrations of protein, fibre and complex carbohydrates. It is an especially important food for many developing countries in Latin America, Asia and Africa (Graham and Vance 2003). It is estimated that the global harvest is ~18.7 million tons and is grown on 27.7 million ha in ~148 countries (Gepts et al. 2008).
Based on the discovery of wild common bean in Argentina (Burkart and Brücher 1953) and Guatemala (McBryde 1947) and archaeological remains found in the Americas (Kaplan and Kaplan 1988; Kaplan and Lynch 1999), common bean is commonly thought to have originated in the Americas. Two large gene pools of wild types were identified based on phaseolin seed protein variation (Gepts et al. 1986; Gepts 1990), DNA marker diversity (Becerra Velasquez and Gepts 1994; Sonnante et al. 1994; Freyre et al. 1996; Tohme et al. 1996), morphology (Evans 1976; Gepts and Debouck 1991), isozymes (Koenig and Gepts 1989), mitochondrial DNA variation (Khairallah et al. 1992) and amplified fragment length polymorphism (AFLP) (Rossi et al. 2009) and short sequence repeats (SSR) (Kwak and Gepts 2009) marker data. The Middle American (MA) gene pool extends from Mexico through Central America and into Venezuela, whereas the Andean gene pool is found in Peru, Chile, Bolivia and Argentina. Recently, multilocus sequence data considered demographic events in wild common bean (S. Mamidi, M. Rossi, D. Annam, S. M. Moghaddam, R. K. Lee, R. Papa, P. E. McClean, unpubl. data) and determined that the two gene pools diverged ~110 000 years before present (BP) followed by a geographical adaptation bottleneck in each wild gene pool.
The discovery of landraces in archeological sites dating from 10 000 years BP in Argentina and 7000 years BP from Mexico (Kaplan et al. 1973) suggests that common bean was domesticated early in Middle and South America. Accelerator mass spectrometry (AMS) analyses provide evidence of cultivation of common bean before ~2500 BP in Tehuacan valley, 1300 BP in Tamaulipas, 2100 BP in Oaxaca and 4400 BP in the Peruvian Andes (Kaplan and Lynch 1999). Linguistic evidence suggests the presence of bean 3400 BP in Middle America (Brown 2006). Mexico is suggested as one centre of domestication of common bean (Gepts et al. 1986; Gepts 1988; Smith 1995; Piperno and Flannery 2001; Doebley 2004) and another centre of domestication is suggested in the Andes (Gepts et al. 1986; Gepts 1998). Apparently, the divergence of ancestral wild common bean at 110 000 years BP provided the genetic basis for the domestication within the two gene pools.
The presence of distinct groups of landraces has been described at both the morphological (Singh et al. 1991a) and molecular level (Gepts et al. 1986; Singh et al. 1991b; Becerra Velasquez and Gepts 1994; Freyre et al. 1996). Singh et al. (1991a) classified the MA cultivars into three races – lowland race Mesoamerican (M) and highland races Durango and Jalisco. Race Durango occupies the semi-arid northern highlands of Mexico. The pinto, great northern, medium red and pink market classes are assigned to this race. Race Jalisco overlaps the southern distribution of the race Durango. Race Mesoamerica, the third MA race, occupies the lowlands of Latin America from Mexico to northern Colombia and Venezuela. Black, navy and small red market classes represent this race. The Andean gene pool is divided into three races based on morphological and ecological criteria – Nueva Granada, Peru and Chile (Singh et al. 1991a). Race Nueva Granada is the most widely cultivated Andean race and includes the majority of the commercial large seeded cultivars. It is grown at mid-altitudes of the Andes and Africa, in warm lowland environments of Brazil, Mexico and the Caribbean and in the temperate climates of North America and Europe. The dark and light red kidney, cranberry and most horticultural snap bean market classes are found within this race. Race Peru is found in the Andean highlands, whereas race Chile occupies the southern Andes.
Although it has been established that domestication was an independent event in each gene pool, the number of domestication events in each gene pool is debated. A single MA domestication event (Gepts et al. 1986; Papa and Gepts 2003; Kwak et al. 2009, Rossi et al. 2009) would imply that divergence into races followed the domestication process instead of resulting from separate domestications. Beebe et al. (2000) used random amplified polymorphic DNA (RAPD) data to suggest that the MA gene pool was the result of two distinct domestication events. This was further supported by chloroplast data (Chacón et al. 2005) and sequence diversity data for DFR and CHI introns (McClean et al. 2004; McClean and Lee 2007).
Archeological and multilocus sequence data provide complementary information to understand the demographic and evolutionary events associated with the domestication of a species (Doebley et al. 2006; Burke et al. 2007). Recent population genomics studies suggest that domestication affects the entire genome and that selection acts on a large number of loci (Wright et al. 2005; Caicedo et al. 2007), so a multilocus approach is appropriate to study the effects of domestication and selection. Similar results were obtained by Papa et al. (2007) on P. vulgaris using 2509 AFLPs. Computationally, approximate Bayesian computation (ABC) has emerged as a preferred approach to simulate many models consisting of various combinations of demographic parameters (some derived from archeological data). Subsequent statistical analyses can then select the model(s) that best fits the observed summary population genetic data obtained from sequencing multiple loci. The ABC simulation method considers the population summary data and make inferences with less computational time than when all available data are analysed in detail. This method was used successfully to untangle many evolutionary processes in humans (Fagundes et al. 2007; Patin et al. 2009; Scheinfeldt et al. 2009; Ray et al. 2010; Batini et al. 2011) and plants (Ross-Ibarra et al. 2007; Ingvarsson 2008; François et al. 2008). The advantages and disadvantages of the methodology are reviewed extensively by Bertorelle et al. (2010), Lopes and Beaumont (2010) and Csilléry et al. (2010). The objectives of this research were to collect multilocus sequence data from domesticated P. vulgaris landraces, evaluate the nucleotide variation within this collection, consider different models of domestication using ABC and estimate the domestication model and population parameters that best describe domestication within each of the two common bean gene pools. The results were compared to previous data at the same loci for wild relatives to assess the genome wide effects of domestication. To our knowledge, this is the first crop species where multiple demographic models were tested to find the best model of domestication.
Materials and methods
Genotypes, genes and DNA sequencing
A collection of 24 landraces were analysed (Table 1). Based on DFR (McClean et al. 2004) and CHI intron-3 (McClean and Lee 2007) haplotypes, unique landraces were selected to represent races within each of the two common gene pools. Phaseolus coccineus L. genotypes PI 325 589 and PI 325 599 were used as out group members. Thirteen nuclear genes were selected for sequencing with at least one locus on each Phaseolus vulgaris (L.) linkage group (Table 2; McConnell et al. 2010). Two additional Pv08 loci, g776 and D1468, were included. D1468 is associated with several domestication traits including number of pods and days to flowering and maturity (Koinange et al. 1996). Locus g776 maps < 2cM from D1468 (McConnell et al. 2010).

|

|
DNA was extracted from young leaves (Brady et al. 1998) and fragments from the 13 loci were amplified using standard PCR conditions. The amplified fragments were sequenced from both directions using a Beckman CEQ 2000XL DNA analysis system (Beckman Coulter Inc., Brea, CA). The DNA sequence chromatograms were analysed using the Staden package (Staden 1996). Gene annotation and structure were determined by blastx against the Viridiplantae database at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed 2 January 2009).
Population genetics statistics
Population structure was determined using the STRUCTURE 2.2 software (Pritchard et al. 2000). Combined data for all loci was used for this analysis. The sequence files were converted into STRUCTURE input format using xmfa2struc (Didelot and Falush 2007). We set k (the number of subpopulations) from 1 to 6 and performed 10 runs for each k value. For each run, a burn in of 100 000 iterations was followed by an additional 500 000 iterations. The Δk method proposed by Evanno et al. (2005) was used to choose the best k value. The assignment of an individual to a subpopulation was based on subpopulation probability values estimated in STRUCTURE. Individuals were assigned to a subpopulation based on a coefficient qi ≥ 0.7. To further differentiate the subpopulations, a neighbour joining (NJ) tree using the combined loci was built in ClustalX (Larkin et al. 2007) and bootstrapped over 1000 replicates.
Unless otherwise stated, population genetic statistics were calculated using DnaSP 4.90 (Rozas and Rozas 1999). Population differentiation was described using Fst (Hudson et al. 1992) and Hudson’s Snn (Hudson 2000) statistic with significance determined by 10 000 permutations. The number of shared (Ss) and fixed (Sf) silent sites between gene pools and the number of unique polymorphic sites (SAnd and SMA) in each population were determined. Nucleotide diversity for synonymous and non-coding silent sites were estimated using (1) the Watterson’s estimator (θw = 4Neµ), (2) the average number of pairwise differences per site between sequences in a sample (π), (3) the number of segregating sites (S), (4) the number of haplotypes (H) and (5) haplotype diversity (Hd). Haplotype diversity is a measure of uniqueness of a haplotype in a population. To test the departure from the neutral equilibrium model of evolution, Tajima’s D (DT; Tajima 1989) was estimated. ZnS, the average R2 (linkage disequilibrium (LD) coefficient) over all pairwise comparisons was also calculated. The expected decay of intergenic LD with physical distance was estimated as described by Remington et al. (2001) and Pyhajarvi et al. (2007) by fitting the data a nonlinear regression equation using the NLIN procedure in SAS 9.2 (SAS Institute, Cary, NC, USA).
Model selection and parameter estimation
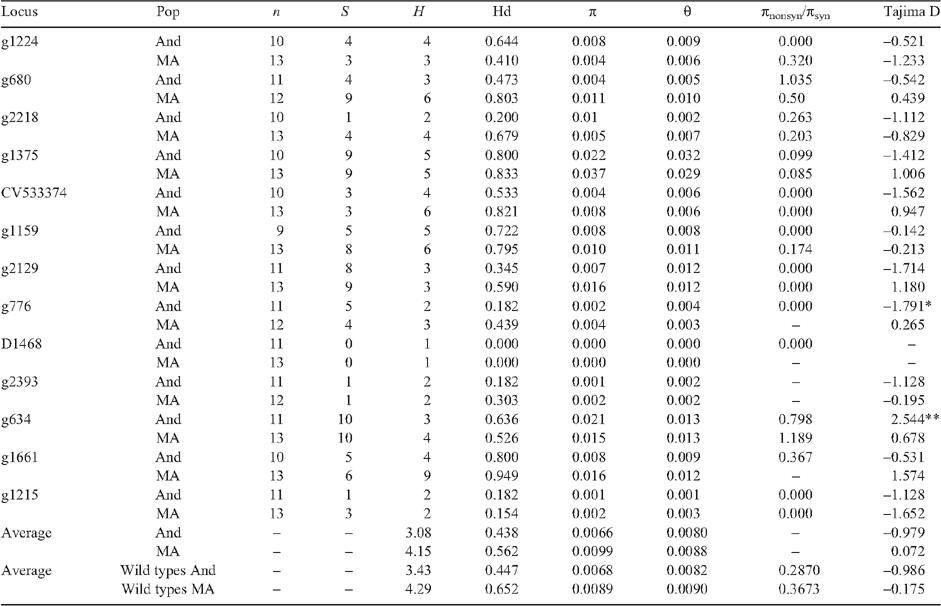
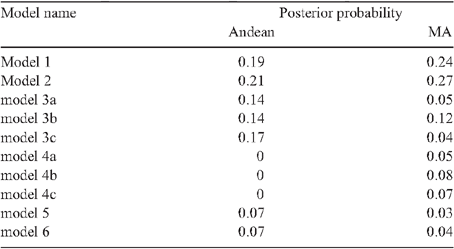
Six domestication models (Fig. 1) were simulated using Hudson’s ms (Hudson 2002) to find the best domestication model in each gene pool. Models 1 and 2 describe a single domestication event in each gene pool. Models 3 and 4 describe the presence of two domestication events, two races together as an event and the other race as another domestication event. Therefore, models 3 and 4 each have three submodels accounting for all three possible combinations in each of the gene pool. Models 5 and 6 indicate the presence of three domestication events, one for each race. All the models consist of a bottleneck during the start of domestication. Models 2, 4 and 6 are characterised by an exponential growth of the landrace population after the bottleneck, whereas models 1, 3 and 5 are characterised by instantaneous expansion of population after the bottleneck. Since population size has little effect on the simulation results (Tenaillon et al. 2004; Wright et al. 2005), ancestral population size (Nw), present population size (NL) and effective population size (Ne) were assumed to be 100 000 individuals. For the MA gene pool models, domestication began at 10 000 years BP and ended before 2500 years BP. For the Andean models, domestication began at 10 000 years BP and ended at 4000 years BP. The ending dates were based on the results by Kaplan and Lynch (1999). A variable mutation rate (µ, based on a uniform distribution of 1 × 10–10 to 1 × 10–6 substitutions per synonymous site per year) and a symmetric migration rate (m, between 1 × 10–2 to 10 individuals per generation) were included in the simulations. The bottleneck population size is assumed to be 0.0001 to 100% of effective population size, equivalent to 1–100 000 individuals. For each locus within the model, we simulated 1 000 000 priors. The simulation results were piped into the msstats 0.2.9 software (available at http://molpopgen.org/) to obtain the summary statistics for each of the simulation. For each simulation, the Euclidean distance between the simulated and observed summary statistics (segregating sites (S), number of haplotypes (H) and nucleotide polymorphism (π)) was calculated. We accepted the top 10 000 simulations for each model with a Euclidean distance <0.1. The best model was selected by combining the accepted simulations across all models and estimating the posterior probability of the model in the top 5000 simulations as described by Pritchard et al. (1999), Estoup et al. (2004) and Ray et al. (2010).
|
For the best model, the summary statistics for the accepted simulations were reduced in dimensionality using principal component analysis (PCA) using PRINCOMP in SAS 9.2. This also helped produce a set of uncorrelated transformed statistics (Ray et al. 2010). Then the parameters were estimated from the accepted 10 000 simulations for the best model using a general linear model (GLM), described by Leuenberger and Wegmann (2010) using the GLM procedure in SAS 9.2. For estimating the goodness of fit of our model and the parameters, we compared the mean of observed statistics with a posterior distribution of summary statistics of the accepted simulations as described by Pascual et al. (2007) and Ingvarsson (2008).
Results
Population genetics statistics
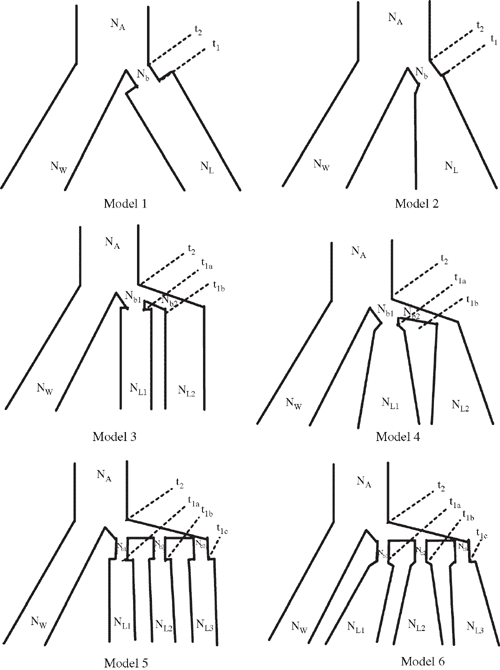
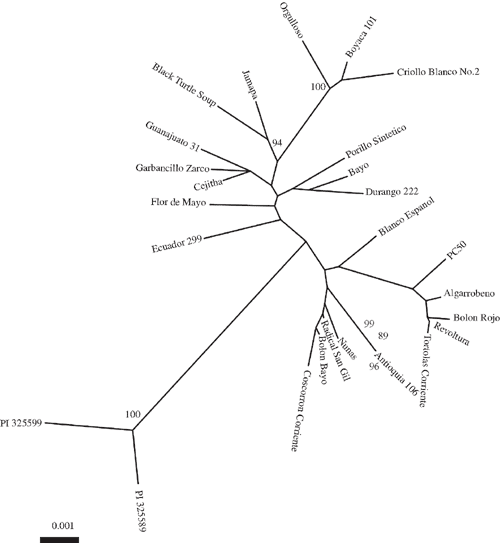
STRUCTURE analysis defined two subpopulations: one composed of Durango, Jalisco and Mesoamerica landraces; and a second represented by Chile, Peru and Nueva Granada landraces. A similar result was observed with a NJ tree where a 100% bootstrap value supported two gene pools (Fig. 2). There was significant genetic differentiation between the two gene pools with an average Fst = 0.38 (Table 3). This was further confirmed with a significant Snn value for 10 loci. The number of shared sites is 38 and there were six fixed sites between the two gene pools. The unique polymorphic sites in MA gene pool (32) were greater than in Andean (18).
|
|
The MA gene pool had 69 segregating sites and the Andean gene pool had 56 segregating sites (Table 4). The average number of MA haplotypes (4.15) and haplotype diversity (0.562) was slightly higher than that of Andean (3.08 and 0.438 respectively). The level of nucleotide diversity was heterogeneous among loci. On average, the polymorphism within landraces was θsil = 0.0088 for MA and θsil = 0.0080 for Andean. For nearly all loci, the ratio of πnonsyn/πsyn was <1 in both gene pools. Based on DT, most loci showed a significant departure from neutral equilibrium expectations in the Andean gene pool (Table 5). DT values were negative for most Andean loci.
|
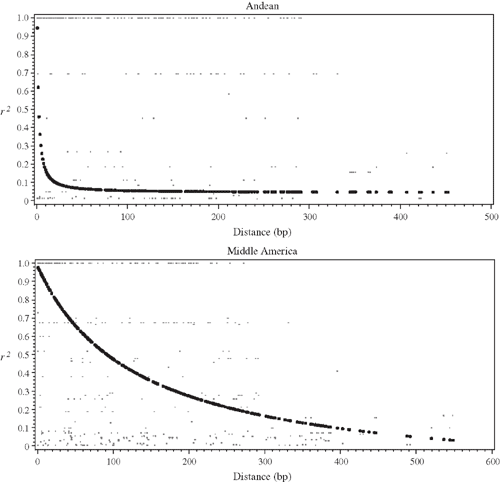
The value of Zns is 0.430 and 0.627 for MA and Andean landraces. In the MA gene pool, LD decayed at 500 bp (r2 < 0.1) whereas in the Andean gene pool, the decay was within 100 bp (r2 = 0.1 (Fig. 3). For the landraces, 42 and 47% of pairwise comparisons were significant in MA and Andean gene pools, respectively, and only 2% were significant in MA and Andean when the Bonferroni correction was applied.
|
Model selection and parameter estimation
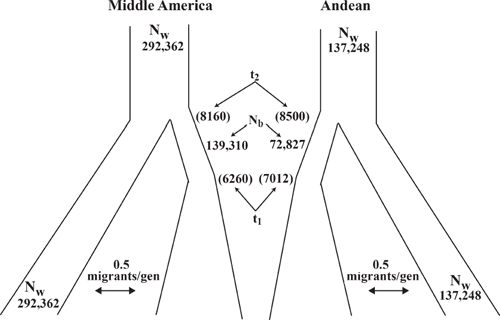
In both gene pools, model 2 performed best with a posterior probability of at least 0.26 in MA and 0.21 in Andean (Table 5). For model 2, seven and three principal components explained 98% of the variability of the summary statistics. For model 2, MA domestication started at 8160 years BP and ended at 6260 years BP with a bottleneck size (in % effective wild types population) of ~48% for the MA gene pool (Table 6; Fig. 4). For model 2 within the Andean gene pool, domestication began 8500 years BP and ended 7012 years BP with a bottleneck size of ~47% (Table 6; Fig. 4). Based on the effective population size estimates of 292 362 and 137 248 for the MA and Andean wild gene pools (S. Mamidi, M. Rossi, D. Annam, S. M. Moghaddam, R. K. Lee, R. Papa, P. E. McClean, unpubl. data), it is estimated that the domestication bottleneck population sizes were 139 310 and 72 827 respectively. The estimated migration rate between the wild and domesticated population in each gene pool was ~0.5 migrants per generation.

|
|
Several simulation summary statistics were compared with the observed mean values to assess the validity of model 2. A reasonable fit was observed for theta (θw) (95% CI = 0–0.324 for MA and 0–0.353 for Andean) and DT (95% CI = 1.451–1.442 for MA and –1.128–0.014 for Andean). The observed means in all cases fell within the 95% distribution of the accepted simulations.
Discussion
Multilocus sequence diversity in common bean
Common bean has a high level of phenotypic diversity illustrated by its wide geographical distribution from northern Mexico to northern Argentina and its adaptation to tropical and temperature environments. Wild types as a whole have the largest level of diversity available (McClean and Lee 2007) and domesticated landraces selected for many important agronomic traits have arisen from the two main wild gene pools. These landraces contain much of the diversity that has been captured in production of cultivars that were mostly developed through hybrid breeding. In this study we evaluated multilocus sequence data from a diverse group of landrace genotypes to assess nucleotide diversity, population differentiation and demography of the species. These results have implications for understanding the history of domestication of common bean, which itself contributes to our understanding of the origin and development of modern cultivation and agronomy (Guo et al. 2010). This study also enhances our understanding of the factors that contribute to LD in present day cultivars which, in turn, has implications for association mapping studies.
The landraces of common bean were divided into two gene-pool specific subpopulations. This observation is consistent with earlier research (Becerra Velasquez and Gepts 1994; Gepts et al. 1986; Singh et al. 1991b; Freyre et al. 1996; McClean et al. 2004; McClean and Lee 2007; Kwak and Gepts 2009). For the landraces we analysed, the nucleotide diversity of MA was higher than that the Andean gene pool, similar to earlier studies (Cattan-Toupance et al. 1998; Beebe et al. 2001; McClean et al. 2004; McClean and Lee 2007; Kwak and Gepts 2009; Rossi et al. 2009). High Fst and significant Snn values and the presence of fixed sites between the two geographically separated gene pools further support their reproductive isolation. The greater haplotype diversity at silent MA sites suggests a larger effective population size for that gene pool.
We found high variance among our DT estimates, with both positive and negative values for the MA subpopulation, which may be due to the influence of evolutionary processes on nucleotide variation (Wright and Gaut 2005; Moeller et al. 2007). Also, this could be due to the initial period of positive DT possible after a bottleneck due to accumulation of intermediate frequency variants (Maruyama and Fuerst 1985; Depaulis et al. 2003). Since the MA gene pool is more diverse than the Andean (Cattan-Toupance et al. 1998; Beebe et al. 2001; Galván et al. 2001; Rossi et al. 2009; Kwak and Gepts 2009), a question for subsequent research is whether selection acted differentially on the MA gene pool that directly influenced neutrality estimates. D1468, a locus previously mapped to a QTL for pod number, days to flowering and maturity (Koinange et al. 1996), had zero diversity in both the gene pools. This may suggest that this locus was selected during the domestication process. However, we do not have the appropriate data to test for the effect of selection at this locus.
The structure analysis of 21 wild types belonging to two gene pools (S. Mamidi, M. Rossi, D. Annam, S. M. Moghaddam, R. K. Lee, R. Papa, P. E. McClean, unpubl. data) along with the landraces under study here suggested the presence of two subpopulations with a probability of assignment of each individual to a group of qi > 0.7. The landraces and wild types of each gene pool were grouped together using STRUCTURE and NJ procedures (data not shown). The close relationship of these is further confirmed by the low Fst values between the wild and landraces within each gene pool (0.15 in MA and 0.04 in Andean). The two major subpopulations identified here are consistent with earlier work of Rossi et al. (2009), Kwak and Gepts (2009) and McClean et al. (2011). This data clearly suggests that landraces within a gene pool arose by a domestication event specific to that gene pool. The hypothesis of independent domestication events in each gene pool was suggested previously (Gepts et al. 1986; Papa and Gepts 2003; Kwak and Gepts 2009; Rossi et al. 2009).
A model for domestication in common bean
Since the domestication event is independent in each gene pool, model selection and parameter estimates were performed separately in each gene pool. Also, since each gene pool is further defined by a specific race structure (Singh et al. 1991a; Beebe et al. 2000), models with 1–3 domestication events were tested using the ABC approach. Even though computationally intensive, this approach has successfully described evolutionary events in many species (Fagundes et al. 2007; Ingvarsson 2008; Patin et al. 2009; Ray et al. 2010; Batini et al. 2011). All models tested included a population bottleneck imposed on a founding population that subsequently led to the landrace population. This is an additional example of another crop species that experienced a bottleneck during domestication (Eyre-Walker et al. 1998, Wright et al. 2005; Zhu et al. 2007). We also evaluated models that included both exponential and instantaneous expansion of the landrace populations. The best models selected in each gene pool consisted of a single domestication event. This single domestication model showed similar posterior probabilities for the two gene pools. These results support hypotheses proposed in common bean by previous researchers (Gepts et al. 1986; Papa and Gepts 2003; Kwak and Gepts 2009; Rossi et al. 2009).
The parameters estimated here suggest a short duration with a large founding population. This result is contrary to other species where the founding population was small and the bottleneck duration was long (Tenaillon et al. 2004; Hamblin et al. 2006; Zhu et al. 2007). Unlike most crops, wild-type common beans belong to the same species and are members of the same gene pool as related landraces. Based on their highly similar phenotypes found for landraces and their wild relatives, along with similar levels of nucleotide diversity, it appears that the domestication bottleneck may have only involved a limited number of genes. The strong population structure of common bean and phenotypic similarity between wild types and landraces could explain the short bottleneck duration.
When wild type and landraces were evaluated together, more shared sites than fixed sites were observed (29 and 0 in MA, 36 and 0 in Andean). This a strong indicator of gene flow and is consistent with the earlier results by Papa and Gepts (2003) and Papa et al. (2005) for common bean, as well the bidirectional gene flow observed in 12 of the 13 most important food crops (Ellstrand et al. 1999; Stewart et al. 2003). Moreover, for 7 of these 13 crops, introgression of domesticated traits increased the competitiveness of the related weed species (Ellstrand et al. 1999; Ellstrand and Schierenbeck 2000). In these cases, gene flow may simply reflect the close proximity of wild types and cultivated landraces in production fields where wild genotypes grow next to cultivated fields in native domains (Papa et al. 2005).
As observed for other plants, a bottleneck increases population structure and reduces within subpopulation diversity. Within each landrace gene pool, population differentiation was higher than that observed for wild types (Fst = 0.29; S. Mamidi, M. Rossi, D. Annam, S. M. Moghaddam, R. K. Lee, R. Papa, P. E. McClean, unpubl. data). This is further supported by presence of more fixed sites in the landraces compared to the wild types (SF = 0; S. Mamidi, M. Rossi, D. Annam, S. M. Moghaddam, R. K. Lee, R. Papa, P. E. McClean, unpubl. data). The higher Fst values for landraces than wild types appear to reflect the cumulative effect of gene flow in wild types over thousands of years, compared to landraces, which are of a much more recent origin. In addition, after the initial domestication event, genotypes became adapted to specific environmental conditions (Kwak and Gepts 2009) and further differentiated into races.
Landrace nucleotide variation was slightly lower than the estimates for the wild types (Table 4). Typically, during domestication, important agronomic characters are selected which result in a genome-wide reduction of genetic diversity in the domesticates (Tanksley and McCouch 1997; Eyre-Walker et al. 1998; Buckler et al. 2001; Diamond 2002; Clark et al. 2004; Papa et al. 2007, Pozzi et al. 2004; Otero-Arnaiz et al. 2005; Vasemagi et al. 2005; Wright et al. 2005; Doebley et al. 2006; Kilian et al. 2007; Zhu et al. 2007). Earlier studies in bean using DNA marker, protein and morphological variation, determined that domesticated landraces indeed contain a subset of the variability found in wild beans (Gepts and Bliss 1986; Gepts et al. 1986; Debouck et al. 1993; Kami et al. 1995; Tohme et al. 1996; Beebe et al. 2001; Chacón et al. 2005). Here we observed that the amount of variability/polymorphisms retained is similar to the diversity estimates found in wild types. These estimates are higher than those proposed by Buckler et al. (2001), who suggested that 60–80% of variability is retained in the domesticated crops. The loss of variability in other crops is within that range: grasses (66%, Buckler et al. 2001), soybean (50%, Hyten et al. 2007; Guo et al. 2010) and rice (70%, Li et al. 2009). In the present study, a high level of loss of diversity is not evident from nucleotide diversity or SNP density data. The SNP density in wild types are one per ~41 bp and ~50 bp for MA and Andean wild types respectively. For landraces, these values are one per ~46 bp and one per ~57 bp. SNP density is higher in other species. For example, wild rice has one SNP per ~19 bp and cultivated rice has one per ~40 bp (Zhu et al. 2007). Similarly there is one SNP per ~19 bp in wild sunflower and one SNP per ~39 bp in cultivated sunflower (Liu and Burke 2006) and one per ~22 bp in teosinte and one per ~30 bp in maize (Tenaillon et al. 2004). This implies that in common bean domestication may have been less severe than in other species. However, we need to consider these results along with other diversity studies using molecular markers (Papa and Gepts 2003; Papa et al. 2007; Kwak et al. 2009; Rossi et al. 2009) where a much larger effect of domestication on the genetic diversity of the common bean was found, particularly in MA genotypes. More analysis at the nucleotide level on larger samples is needed in order to better estimate the selection intensity and the drift associated with domestication in common bean.
In general, it would be expected that LD for self-pollinated species like P. vulgaris would extend to the kilobase level as observed for Arabidopsis thaliana (L.) Heynh (Nordborg et al. 2002), rice (Garris et al. 2003) and soybean (Zhu et al. 2003). The lack of LD decay in Andean landraces is reflective of the low diversity levels and low population differentiation within the gene pool. The mean LD coefficient (Zns) estimate is higher for Andean landraces than MA landraces, which is consistent with earlier results by Rossi et al. (2009).
A higher level of LD in landraces than wild populations is likely due to lower diversity and the short time frame to accumulate recombination events among the domesticated genotypes (Morrell et al. 2005; Caldwell et al. 2006; Rostoks et al. 2006; Hyten et al. 2007). The higher level of LD in Andean compared to MA gene pool in both wild types (0.27 in MA and 0.46 in Andean) and landraces suggests that the higher LD in the Andean gene pool originated before domestication (Rossi et al. 2009) and is suggested to be the result of migration, genetic drift and selection (Rossi et al. 2009). LD decay in landraces is more than that of wild types, consistent with previous estimates that suggested increase in LD decay distance after a bottleneck (Flint-Garcia et al. 2003; Gupta et al. 2005; Li et al. 2009). Similar estimates have been observed in other crops (Morrell et al. 2005; Caldwell et al. 2006; Liu and Burke 2006; Zhu et al. 2007).
Wild common bean is divided into two gene pools (Singh et al. 1991a, 1991b; Blair et al. 2006; Díaz and Blair 2006; McClean et al. 2004; McClean and Lee 2007; Kwak and Gepts 2009; Rossi et al. 2009) that appear to have arisen from a common ancestor 110 000 years BP (S. Mamidi, M. Rossi, D. Annam, S. M. Moghaddam, R. K. Lee, R. Papa, P. E. McClean, unpubl. data). Both wild gene pools arose via a bottleneck probably associated with regional adaptation. Only one domestication event is suggested in each landrace gene pool that is consistent with suggestions from previous research (Kwak and Gepts 2009; Rossi et al. 2009). The domestication events in each gene pool were characterised by a bottleneck of ~50% effective population size and a bottleneck length of ~2000 and 1500 years in the MA and Andean gene pools respectively. After the bottleneck, diversification of landraces into races occurred which differ by morphological and physiological characteristics.
Effects of domestication history on association mapping in common bean
Association mapping uses the linkage disequilibrium in a population of choice to discover QTL for various traits of importance. The major advantage of association mapping vs bi-parental mapping is that it samples more recombination events are available than in a single pair-wise cross. This presumably will lead to higher mapping resolution (Myles et al. 2009). As we discuss below, the results here have important implications for association mapping in common bean.
Different demographic factors influence LD in different ways. Bottlenecks reduce genetic variation and change the gene frequency spectrum by removing low-frequency alleles (Hamblin et al. 2011). The extent of LD increases due to the elimination of a subset of recombination events. Selection also increases LD distance that may extend beyond the average for the whole genome (Myles et al. 2009). These factors can lead to an extensive haplotype structure which is more pronounced in self-pollinating crops (Hamblin et al. 2011). In P. vulgaris, the two gene pools diverged, with a reduction in diversity due to bottlenecks, in wild types at 110 000 years BP. The domestication bottleneck within each gene pool, the subsequent differentiation into races and selection by breeders to develop cultivars led to an increase in LD. Finally, as a result of the its self-pollinating nature, it is difficult to break up the LD generated by these factors in common bean (Myles et al. 2009; Hamblin et al. 2011) and mapping resolution would generally be low. A final concern is the high degree of population structure observed in common bean. As has been documented, population structure can result in spurious associations (false positives) between phenotypes and unlinked markers (Knowler et al. 1988; Cardon and Palmer 2003). Even though mixed linear models, which account for population structure and relatedness can minimise the discovery of false positives, this is at the expense of reducing the power to detect true positives (Zhao et al. 2007; Brachi et al. 2010).
To counteract these effects, we propose that common bean association mapping should be performed with populations consisting of individuals from within a single gene pool. First, this will greatly reduce the population structure problem often observed for common bean populations (Kwak and Gepts 2009; Rossi et al. 2009; McClean et al. 2011). This will be most beneficial for mapping in the Andean gene pool because the very low genetic differentiation among the races and the number of markers required would be high due to its low LD decay. Depending on the specific population selected, it might be possible, as with humans, to perform the mapping without correcting for population structure. As for mapping in the MA gene pool, it may even be of further benefit to consider populations derived from the Durango and Jalisco races as a pool and Mesoamerican races as a second pool. Population structure analyses consistently define Durango and Jalisco landraces as a single subpopulation and Mesoamerican genotypes as a second subpopulation (McClean et al. 2011). Due to a significant LD decay distance, fewer markers would be necessary. Given the low diversity within each of these subpopulations, it will be necessary to use a much larger core set of SNP markers to discover those polymorphic within these distinct subpopulations. Soon this should not be a concern, given the low cost of discovering SNPs using massively parallel next generation techniques.
References
Batini C, Lopes J, Behar DM, Calafell F, Jorde LB, van der Veen L, Quintana-Murci L, Spedini G, Destro-Bisol G, Comas D (2011) Insights into the demographic history of African pygmies from complete mitochondrial genomes. Molecular Biology and Evolution 28, 1099–1110.| Insights into the demographic history of African pygmies from complete mitochondrial genomes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXnsVSrtw%3D%3D&md5=ed0e3531a4c54e3bbae9e6839cb129d9CAS |
Becerra Velasquez VL, Gepts P (1994) RFLP diversity of common bean (Phaseolus vulgaris) in its centers of origin. Genome 37, 256–263.
| RFLP diversity of common bean (Phaseolus vulgaris) in its centers of origin.Crossref | GoogleScholarGoogle Scholar |
Beebe S, Skroch PW, Tohme J, Duque MC, Pedraza F, Nienhuis J (2000) Structure of genetic diversity among common bean landraces of Middle American origin based on correspondence analysis of RAPD. Crop Science 40, 264–273.
| Structure of genetic diversity among common bean landraces of Middle American origin based on correspondence analysis of RAPD.Crossref | GoogleScholarGoogle Scholar |
Beebe S, Rengifo J, Gaitan E, Duque MC, Tohme J (2001) Diversity and origin of Andean landraces of common bean. Crop Science 41, 854–862.
| Diversity and origin of Andean landraces of common bean.Crossref | GoogleScholarGoogle Scholar |
Bertorelle G, Benazzo A, Mona S (2010) ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Molecular Ecology 19, 2609–2625.
| ABC as a flexible framework to estimate demography over space and time: some cons, many pros.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3cnnvVKgug%3D%3D&md5=4618014a82d4d243f5c23473dd43b410CAS |
Blair MW, Iriarte G, Beebe S (2006) QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean × wild common bean (Phaseolus vulgaris L.) cross. Theoretical and Applied Genetics 112, 1149–1163.
| QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean × wild common bean (Phaseolus vulgaris L.) cross.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjtVSmtrY%3D&md5=8a82958b76059e2f8b63963e070f101bCAS |
Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F (2010) Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLOS Genetics 6, e1000940
| Linkage and association mapping of Arabidopsis thaliana flowering time in nature.Crossref | GoogleScholarGoogle Scholar |
Brady L, Bassett MJ, McClean PE (1998) Molecular markers associated with T and Z, two genes controlling partly coloured seed coat patterns in common bean. Crop Science 38, 1073–1075.
| Molecular markers associated with T and Z, two genes controlling partly coloured seed coat patterns in common bean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXlvFWktrY%3D&md5=06cd4c8aa6fc53ee22193fe85b5de879CAS |
Brown CH (2006) Prehistoric chronology of the common bean in the New World: the linguistic evidence. American Anthropologist 108, 507–516.
| Prehistoric chronology of the common bean in the New World: the linguistic evidence.Crossref | GoogleScholarGoogle Scholar |
Buckler ES, Thornsberry JM, Kresovich S (2001) Molecular diversity, structure and domestication of grasses. Genetical Research 77, 213–218.
| Molecular diversity, structure and domestication of grasses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXmtVKit7c%3D&md5=d0791bf143dcf0c025c7852d281f2bd0CAS |
Burkart A, Brücher H (1953) Phaseolus aborigineus Burkart, die mutmassliche andine Stammform der Kulturbohne. Züchter 23, 65–72.
Burke JM, Burger JC, Chapman MA (2007) Crop evolution: from genetics to genomics. Current Opinion in Genetics & Development 17, 525–532.
| Crop evolution: from genetics to genomics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhsVWisbzL&md5=4a5fd64b99358806764da649ffdf4795CAS |
Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR, Bustamante CD, Purugganan MD (2007) Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLOS Genetics 3, e163
| Genome-wide patterns of nucleotide polymorphism in domesticated rice.Crossref | GoogleScholarGoogle Scholar |
Caldwell KS, Russell J, Langridge P, Powell W (2006) Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172, 557–567.
| Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhvV2jt7s%3D&md5=69a231ea73151a18ab7a699499435165CAS |
Cardon LR, Palmer LJ (2003) Population structure and spurious associations. Lancet 361, 598–604.
| Population structure and spurious associations.Crossref | GoogleScholarGoogle Scholar |
Cattan-Toupance I, Michalakis Y, Neema C (1998) Genetic structure of wild bean populations in their South-Andean centre of origin. Theoretical and Applied Genetics 96, 844–851.
| Genetic structure of wild bean populations in their South-Andean centre of origin.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXksFSiu74%3D&md5=693d70315fc72208df0fc49c14a27a7cCAS |
Chacón S, Pickersgill B, Debouck DG (2005) Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of the Mesoamerican and Andean cultivated races. Theoretical and Applied Genetics 110, 432–444.
| Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of the Mesoamerican and Andean cultivated races.Crossref | GoogleScholarGoogle Scholar |
Clark RM, Linton E, Messing J, Doebley JF (2004) Pattern of diversity in the genomic region near the maize domestication gene tb1. Proceedings of the National Academy of Sciences of the United States of America 101, 700–707.
| Pattern of diversity in the genomic region near the maize domestication gene tb1.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhsVWhtbo%3D&md5=920c6fb1fb5cd4141a4d485c932f9ee6CAS |
Csilléry K, Blum MGB, Gaggiotti OE, François O (2010) Approximate Bayesian computation (ABC) in practice. Trends in Ecology & Evolution 25, 410–418.
| Approximate Bayesian computation (ABC) in practice.Crossref | GoogleScholarGoogle Scholar |
Debouck DG, Toro O, Paredes OM, Johnson WC, Gepts P (1993) Genetic diversity and ecological distribution of Phaseolus vulgaris (Fabaceae) in northwestern South America. Economic Botany 47, 408–423.
| Genetic diversity and ecological distribution of Phaseolus vulgaris (Fabaceae) in northwestern South America.Crossref | GoogleScholarGoogle Scholar |
Depaulis F, Mousset S, Veuille M (2003) Power of neutrality tests to detect bottlenecks and hitchhiking. Journal of Molecular Evolution 57, S190–S200.
| Power of neutrality tests to detect bottlenecks and hitchhiking.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjsVeiuw%3D%3D&md5=0af1bfbfa8dd1d3bcbdec65ab4c0adafCAS |
Diamond J (2002) Evolution, consequences and future of plant and animal domestication. Nature 418, 700–707.
| Evolution, consequences and future of plant and animal domestication.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XlvVyltbY%3D&md5=e97ace3dbf7feb36b4c01a382c443227CAS |
Díaz LM, Blair MW (2006) Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers. Theoretical and Applied Genetics 114, 143–154.
| Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers.Crossref | GoogleScholarGoogle Scholar |
Didelot X, Falush D (2007) Inference of bacterial microevolution using multilocus sequence data. Genetics 175, 1251–1266.
| Inference of bacterial microevolution using multilocus sequence data.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXltFKhs7k%3D&md5=8bdb64fbcb83dff0e647dc8f8b9a34beCAS |
Doebley J (2004) The genetics of maize evolution. Annual Review of Genetics 38, 37–59.
| The genetics of maize evolution.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXltlOqsg%3D%3D&md5=3644b31daeeeb91dc5c0cacf2a185f72CAS |
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127, 1309–1321.
| The molecular genetics of crop domestication.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmvVyluw%3D%3D&md5=b7feb0b18fa40f4c318b72526193b847CAS |
Ellstrand NC, Schierenbeck KA (2000) Hybridisation as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the United States of America 97, 7043–7050.
| Hybridisation as a stimulus for the evolution of invasiveness in plants?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXksVKisbk%3D&md5=fc14ea83c28d05277214c9cc100cc927CAS |
Ellstrand NC, Prentice HC, Hancock JF (1999) Gene flow and introgression from domesticated plants into their wild relatives. Annual Review of Ecology Evolution and Systematics 30, 539–563.
| Gene flow and introgression from domesticated plants into their wild relatives.Crossref | GoogleScholarGoogle Scholar |
Estoup A, Beaumont M, Sennedot F, Moritz C, Cornuet JM (2004) Genetic analysis of complex demographic scenarios: spatially expanding populations of the cane toad, Bufo marinus. Evolution 58, 2021–2036.
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14, 2611–2620.
| Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmvF2qtrg%3D&md5=5aede1df804e28ef81e7c9a92a5b45afCAS |
Evans A (1976) Beans. In ‘Evolution of crops plants’. (Ed. J Smartt, NW Simmonds) pp. 168–172. (Longman: London)
Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS (1998) Investigation of the bottleneck leading to the domestication of maize. Proceedings of the National Academy of Sciences of the United States of America 95, 4441–4446.
| Investigation of the bottleneck leading to the domestication of maize.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXis1Ogsr4%3D&md5=28432e0a918cff99abd26282b55f5602CAS |
Fagundes NJR, Ray N, Beaumont M, Neuenschwander S, Salzano FM, Bonatto SL, Excoffier L (2007) Statistical evaluation of alternative models of human evolution. Proceedings of the National Academy of Sciences of the United States of America 104, 17614–17619.
| Statistical evaluation of alternative models of human evolution.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXht12msbjL&md5=6a727f05b1223ef5dfc3a4cc25836378CAS |
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annual Review of Plant Biology 54, 357–374.
| Structure of linkage disequilibrium in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXntFSgu7c%3D&md5=fa18b79c7b1d5cb4cc6db1a108465bbdCAS |
François O, Blum MGB, Jakobsson M, Rosenberg NA (2008) Demographic history of European populations of Arabidopsis thaliana. PLOS Genetics 4, e1000075
| Demographic history of European populations of Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar |
Freyre R, Ríos R, Guzmán L, Debouck DG, Gepts P (1996) Ecogeographic distribution of Phaseolus spp. (Fabaceae) in Bolivia. Economic Botany 50, 195–215.
| Ecogeographic distribution of Phaseolus spp. (Fabaceae) in Bolivia.Crossref | GoogleScholarGoogle Scholar |
Fuller DQ (2007) Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the Old World. Annals of Botany 100, 903–924.
| Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the Old World.Crossref | GoogleScholarGoogle Scholar |
Galván MZ, Aulicino MB, García Medina S, Balatti PA (2001) Genetic diversity among Northwestern Argentinian cultivars of common bean (Phaseolus vulgaris L.) as revealed by RAPD markers. Genetic Resources and Crop Evolution 48, 251–260.
| Genetic diversity among Northwestern Argentinian cultivars of common bean (Phaseolus vulgaris L.) as revealed by RAPD markers.Crossref | GoogleScholarGoogle Scholar |
Garris AJ, McCouch SR, Kresovich S (2003) Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165, 759–769.
Gepts P (1988) Phaseolin as an evolutionary marker. In ‘Genetic resources of Phaseolus beans’. (Ed. P Gepts) pp. 215–241. (Kluwer Academic Publishers: Dordrecht, The Netherlands)
Gepts P (1990) Biochemical evidence bearing on the domestication of Phaseolus (Fabaceae) beans. Economic Botany 44, 28–38.
| Biochemical evidence bearing on the domestication of Phaseolus (Fabaceae) beans.Crossref | GoogleScholarGoogle Scholar |
Gepts P (1998) Origin and evolution of common bean: past events and recent trends. HortScience 33, 1124–1130.
Gepts P, Bliss FA (1986) Phaseolin variability among wild and cultivated common beans (Phaseolus vulgaris) from Colombia. Economic Botany 40, 469–478.
| Phaseolin variability among wild and cultivated common beans (Phaseolus vulgaris) from Colombia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2sXhsFyksQ%3D%3D&md5=b83bcb14adfe0abd5ee0b4762a41ae06CAS |
Gepts P, Debouck D (1991) Origin, domestication and evolution of the common bean (Phaseolus vulgaris L.). In ‘Common beans: research for crop improvement’. (Eds A Van Schoonhoven, O Voysest) pp. 7–53. (CAB International: Wallingford, UK)
Gepts P, Osborn TC, Rashka K, Bliss FA (1986) Phaseolin-protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris): evidence for multiple centers of domestication. Economic Botany 40, 451–468.
| Phaseolin-protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris): evidence for multiple centers of domestication.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2sXhsFyksA%3D%3D&md5=414bdf1a96212cd3d881dde43030f4a8CAS |
Gepts P, Aragão FJL, de Barros E, Blair MW, Brondani R, Broughton W, Galasso I, Hernández G, Kami J, Lariguet P, McClean P, Melotto M, Miklas P, Pauls P, Pedrosa-Harand A, Porch T, Sánchez F, Sparvoli F, Yu K (2008) Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the tropics. In ‘Genomics of tropical crop plants’. (Ed. PH Moore, R Ming) pp. 113–143. (Springer: New York)
Glémin S, Bataillon T (2009) A comparative view of the evolution of grasses under domestication. New Phytologist 183, 273–290.
| A comparative view of the evolution of grasses under domestication.Crossref | GoogleScholarGoogle Scholar |
Graham P, Vance C (2003) Legumes: importance and constraints to greater use. Plant Physiology 131, 872–877.
| Legumes: importance and constraints to greater use.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXisFemtb4%3D&md5=3ff6999509d179d807ee0c3cd336fa4bCAS |
Guo J, Wang Y, Song C, Zhou J, Qiu L, Huang H, Wang Y (2010) A single origin and moderate bottleneck during domestication of soybean (Glycine max): implications from microsatellites and nucleotide sequences. Annals of Botany 106, 505–514.
| A single origin and moderate bottleneck during domestication of soybean (Glycine max): implications from microsatellites and nucleotide sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtVGru7%2FK&md5=e27c563eee563818a0abde5b0867739aCAS |
Gupta PK, Rustgi S, Kulwal PL (2005) Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Molecular Biology 57, 461–485.
| Linkage disequilibrium and association studies in higher plants: present status and future prospects.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjtV2gsrk%3D&md5=6058fa6875f77d5135718d6bb2caf542CAS |
Hamblin MT, Casa AM, Sun H, Murray SC, Paterson AH, Aquadro CF, Kresovich S (2006) Challenges of detecting directional selection after a bottleneck: lessons from Sorghum bicolor. Genetics 173, 953–964.
| Challenges of detecting directional selection after a bottleneck: lessons from Sorghum bicolor.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xns12qsb4%3D&md5=be00034bd6d4845e5f739176bf21e4c3CAS |
Hamblin MT, Buckler ES, Jannink JL (2011) Population genetics of genomics-based crop improvement methods. Trends in Genetics 27, 98–106.
| Population genetics of genomics-based crop improvement methods.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjsV2htro%3D&md5=c9925aef5b32994d8feda9bcfb2f9113CAS |
Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155, 2011–2014.
Hudson RR (2002) Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18, 337–338.
| Generating samples under a Wright–Fisher neutral model of genetic variation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XitFentrw%3D&md5=fbec188edf66396b811481f03aa6898bCAS |
Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589.
Hyten DL, Choi IY, Song QJ, Shoemaker RC, Nelson RL, Costa JM, Specht JE, Cregan PB (2007) Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics 175, 1937–1944.
| Highly variable patterns of linkage disequilibrium in multiple soybean populations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmsFylsL4%3D&md5=ddd92a4913b5e6446a6c746dd7e78036CAS |
Ingvarsson P (2008) Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics 180, 329–340.
| Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula.Crossref | GoogleScholarGoogle Scholar |
Kami J, Velásquez VB, Debouck DG, Gepts P (1995) Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proceedings of the National Academy of Sciences of the United States of America 92, 1101–1104.
| Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXjslegs7w%3D&md5=5529c4e36d62304aba88a2fc2604b123CAS |
Kaplan L, Kaplan L (1988) Genetic resources of Phaseolus beans: their maintenance, domestication, evolution and utilisation. In ‘Current plant science and biotechnology in agriculture’. (Ed. P Gepts) pp. 125–142. (Kluwer Academic Publishers: Dordrecht, The Netherlands)
Kaplan L, Lynch TF (1999) Phaseolus (Fabaceae) in archaeology: AMS. Economic Botany 53, 261–272.
| Phaseolus (Fabaceae) in archaeology: AMS.Crossref | GoogleScholarGoogle Scholar |
Kaplan L, Lynch TF, Smith CE (1973) Early cultivated beans (Phaseolus vulgaris) from an intermontane Peruvian valley. Science 179, 76–77.
| Early cultivated beans (Phaseolus vulgaris) from an intermontane Peruvian valley.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3czpslGktg%3D%3D&md5=f5b8d0e47592b8bad85e18bac01cafc2CAS |
Khairallah MM, Sears BB, Adams MW (1992) Mitochondrial restriction fragment length polymorphisms in wild Phaseolus vulgaris L.: insights on the domestication of the common bean. Theoretical and Applied Genetics 84, 915–922.
| Mitochondrial restriction fragment length polymorphisms in wild Phaseolus vulgaris L.: insights on the domestication of the common bean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXnsVGntg%3D%3D&md5=d99ec7e6180ea21772c7604e3b54c94cCAS |
Kilian B, Ozkan H, Walther A, Kohl J, Dagan T, Salamini F, Martin W (2007) Molecular diversity at 18 loci in 321 wild and 92 domesticate lines reveal no reduction of nucleotide diversity during Triticum monococcum (Einkorn) domestication: implications for the origin of agriculture. Molecular Biology and Evolution 24, 2657–2668.
| Molecular diversity at 18 loci in 321 wild and 92 domesticate lines reveal no reduction of nucleotide diversity during Triticum monococcum (Einkorn) domestication: implications for the origin of agriculture.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXnsVGjuw%3D%3D&md5=5ee0e68b68248e2a65e0cd3efc6ea229CAS |
Knowler WC, Williams RC, Pettitt DJ, Steinberg AG (1988) Gm3;5,13,14 and type 2 diabetes mellitus: an association in American Indians with genetic admixture. American Journal of Human Genetics 43, 520–526.
Koenig R, Gepts P (1989) Allozyme diversity in wild Phaseolus vulgaris: further evidence for two major centers of genetic diversity. Theoretical and Applied Genetics 78, 809–817.
| Allozyme diversity in wild Phaseolus vulgaris: further evidence for two major centers of genetic diversity.Crossref | GoogleScholarGoogle Scholar |
Koinange EMK, Singh SP, Gepts P (1996) Genetic control of the domestication syndrome in common bean. Crop Science 36, 1037–1044.
| Genetic control of the domestication syndrome in common bean.Crossref | GoogleScholarGoogle Scholar |
Kwak M, Gepts P (2009) Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theoretical and Applied Genetics 118, 979–992.
| Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXis12gsLo%3D&md5=79c951371d3080fa3cd3e0c9ac821874CAS |
Kwak M, Kami JA, Gepts P (2009) The putative Mesoamerican domestication center of Phaseolus vulgaris is located in the Lerma-Santiago basin of Mexico. Crop Science 49, 554–563.
| The putative Mesoamerican domestication center of Phaseolus vulgaris is located in the Lerma-Santiago basin of Mexico.Crossref | GoogleScholarGoogle Scholar |
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
| Clustal W and Clustal X version 2.0.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtlaqsL%2FM&md5=e14681fe79aeb2676112db754dffc0f6CAS |
Leuenberger C, Wegmann D (2010) Bayesian computation and model selection without likelihoods. Genetics 184, 243–252.
| Bayesian computation and model selection without likelihoods.Crossref | GoogleScholarGoogle Scholar |
Li X, Tan L, Zhu Z, Huang H, Liu Y, Hu S, Sun C (2009) Patterns of nucleotide diversity in wild and cultivated rice. Plant Systematics and Evolution 281, 97–106.
| Patterns of nucleotide diversity in wild and cultivated rice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXoslalu7c%3D&md5=f5684956bf848627c6f70aa2136cbd5cCAS |
Liu A, Burke JM (2006) Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics 173, 321–330.
| Patterns of nucleotide diversity in wild and cultivated sunflower.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XlvF2itrw%3D&md5=cc0e92d9cf2fe4a1ed6a6739a7b87015CAS |
Lopes JS, Beaumont ME (2010) ABC: a useful Bayesian tool for the analysis of population data. Infection, Genetics and Evolution 10, 825–832.
| ABC: a useful Bayesian tool for the analysis of population data.Crossref | GoogleScholarGoogle Scholar |
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111, 675–689.
McBryde F (1947) Cultural and historical geography of southwest Guatemala. Smithsonian Institute Publication 4, 1–184.
McClean PE, Lee RK (2007) Genetic architecture of chalcone isomerase non-coding regions in common bean (Phaseolus vulgaris L.). Genome 50, 203–214.
| Genetic architecture of chalcone isomerase non-coding regions in common bean (Phaseolus vulgaris L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXntlOguro%3D&md5=93bc4e1f82fb734cccf8dde332ffcf54CAS |
McClean P, Lee R, Miklas P (2004) Sequence diversity analysis of dihydroflavonol 4-reductase intron 1 in common bean. Genome 47, 266–280.
| Sequence diversity analysis of dihydroflavonol 4-reductase intron 1 in common bean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXksFOrs7Y%3D&md5=a6a9b93593d7c6fef1ec7b5ccb290b71CAS |
McClean PE, Terpstra J, McConnell M, White C, Lee R, Mamidi S (2011) Population structure and genetic differentiation among the USDA common bean (Phaseolus vulgaris L.) core collection. Genetic Resources and Crop Evolution in press.
McConnell M, Mamidi S, Lee R, Chikara S, Rossi M, Papa R, McClean P (2010) Syntenic relationships among legumes revealed using a gene-based genetic linkage map of common bean (Phaseolus vulgaris L.) Theoretical and Applied Genetics 121, 1103–1116.
| Syntenic relationships among legumes revealed using a gene-based genetic linkage map of common bean (Phaseolus vulgaris L.)Crossref | GoogleScholarGoogle Scholar |
Moeller DA, Tenaillon MI, Tiffin P (2007) Population structure and its effects on patterns of nucleotide polymorphism in teosinte (Zea mays ssp. parviglumis). Genetics 176, 1799–1809.
| Population structure and its effects on patterns of nucleotide polymorphism in teosinte (Zea mays ssp. parviglumis).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtVeiur3P&md5=df432f8a512bc01b2ac4920d3bc2115bCAS |
Morrell PL, Toleno DM, Lundy KE, Clegg MT (2005) Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilisation. Proceedings of the National Academy of Sciences of the United States of America 102, 2442–2447.
| Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilisation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhvFKitLk%3D&md5=91aec95315c37c5aa3f0affa4e4244efCAS |
Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell Online 21, 2194–2202.
| Association mapping: critical considerations shift from genotyping to experimental design.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXht12qsL3P&md5=a3136696d920edf94476f6a5d5d2b662CAS |
Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloff JN, Noyes T, Oefner PJ, Stahl EA, Weigel D (2002) The extent of linkage disequilibrium in Arabidopsis thaliana. Nature Genetics 30, 190–193.
| The extent of linkage disequilibrium in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XhtFWltLo%3D&md5=db47699745f432c49bac458fa890720fCAS |
Otero-Arnaiz A, Casas A, Hamrick JL, Cruse-Sanders J (2005) Genetic variation and evolution of Polaskia chichipe (Cactaceae) under domestication in the Tehucan Valley, central Mexico. Molecular Ecology 14, 1603–1611.
| Genetic variation and evolution of Polaskia chichipe (Cactaceae) under domestication in the Tehucan Valley, central Mexico.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXkvVeqsbo%3D&md5=ae0abf7671d0a94ef79382d62b087749CAS |
Papa R, Gepts P (2003) Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theoretical and Applied Genetics 106, 239–250.
Papa R, Acosta J, Delgado-Salinas A, Gepts P (2005) A genome-wide analysis of differentiation between wild and domesticated Phaseolus vulgaris from Mesoamerica. Theoretical and Applied Genetics 111, 1147–1158.
| A genome-wide analysis of differentiation between wild and domesticated Phaseolus vulgaris from Mesoamerica.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtV2ksbjL&md5=1fb912c49048e7a0f2be2f6d9dac3c16CAS |
Papa R, Bellucci E, Rossi M, Leonardi S, Rau D, Gepts P, Nanni L, Attene G (2007) Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples. Annals of Botany 100, 1039–1051.
| Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXivVartg%3D%3D&md5=f8787d665e4ed2d40e67d4e2695f4b43CAS |
Pascual M, Chapuis MP, Mestres F, Balanyà J, Huey RB, Gilchrist GW, Serra L, Estoup A (2007) Introduction history of Drosophila subobscurain the New World: a microsatellite based survey using ABC methods. Molecular Ecology 16, 3069–3083.
| Introduction history of Drosophila subobscurain the New World: a microsatellite based survey using ABC methods.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtVertrfK&md5=93af63e959f5b968a439918e50563f84CAS |
Patin E, Laval G, Barreiro LB, Salas A, Semino O, Santachiara-Benerecetti S, Kidd KK, Kidd JR, Van der Veen L, Hombert JM, Gessain A, Froment A, Bahuchet S, Heyer E, Quintana-Murci L (2009) Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLOS Genetics 5, e1000448
| Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set.Crossref | GoogleScholarGoogle Scholar |
Piperno DR, Flannery KV (2001) The earliest archaeological maize (Zea mays L.) from highland Mexico: new accelerator mass spectrometry dates and their implications. Proceedings of the National Academy of Sciences of the United States of America 98, 2101–2103.
| The earliest archaeological maize (Zea mays L.) from highland Mexico: new accelerator mass spectrometry dates and their implications.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXhsVWitLk%3D&md5=40f000dce0795faa669a7bce89f1936bCAS |
Pozzi C, Rossini L, Vecchietti A, Salamini F (2004) Gene and genome changes during domestication. In ‘Cereal genomics’. (Eds PK Gupta, RK Varshney) pp. 165–198. (Kluwier Academic Publishers: London)
Pritchard JK, Seielstad MT, Perez-Lezaun A, Feldman MW (1999) Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Molecular Biology and Evolution 16, 1791–1798.
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Pyhajarvi T, Garcia-Gil MR, Knurr T, Mikkonen M, Wachowiak W, Savolainen O (2007) Demographic history has influenced nucleotide diversity in European Pinus sylvestris populations. Genetics 177, 1713–1724.
| Demographic history has influenced nucleotide diversity in European Pinus sylvestris populations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXivFSrtg%3D%3D&md5=985ea9810f980295bdc5848a51b9e0b7CAS |
Ray N, Wegmann D, Fagundes NJR, Wang S, Ruiz-Linares A, Excoffier L (2010) A statistical evaluation of models for the initial settlement of the American continent emphasizes the importance of gene flow with Asia. Molecular Biology and Evolution 27, 337–345.
| A statistical evaluation of models for the initial settlement of the American continent emphasizes the importance of gene flow with Asia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXntlGnug%3D%3D&md5=2d8afe08fd09d420bd4566c73f5bd7a9CAS |
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Sciences of the United States of America 98, 11479–11484.
| Structure of linkage disequilibrium and phenotypic associations in the maize genome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXnt1yqu7s%3D&md5=a8fdda95b8c7e3327876d2dd5d6e93a2CAS |
Ross-Ibarra J, Morrell PL, Gaut BS (2007) Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proceedings of the National Academy of Sciences of the United States of America 104, 8641–8648.
| Plant domestication, a unique opportunity to identify the genetic basis of adaptation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXlvFCqu7k%3D&md5=00ec0b81969e56e3ca9bc715dc255069CAS |
Rossi M, Bitocchi E, Bellucci E, Nanni L, Rau D, Attene G, Papa R (2009) Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evolutionary Applications 2, 504–522.
| Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L.Crossref | GoogleScholarGoogle Scholar |
Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML, Svensson JT, Stein N, Varshney RK, Marshall DF, Graner A, Close TJ, Waugh R (2006) Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proceedings of the National Academy of Sciences of the United States of America 103, 18656–18661.
| Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtlahtrfP&md5=c7fac75f07a3a4ccbda3d1ccdeb6cf0aCAS |
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15, 174–175.
| DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXisVOksrY%3D&md5=b05aa94b4a1cba8ac81ff8311e4d530aCAS |
Scheinfeldt LB, Biswas S, Madeoy J, Connelly CF, Schadt EE, Akey JM (2009) Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history. Molecular Biology and Evolution 26, 1357–1367.
| Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmtFSnt7k%3D&md5=11dfc6b45579c4fc0854206538f85e9bCAS |
Singh SP, Gepts P, Debouck DG (1991a) Races of common bean (Phaseolus vulgaris, Fabaceae). Economic Botany 45, 379–396.
| Races of common bean (Phaseolus vulgaris, Fabaceae).Crossref | GoogleScholarGoogle Scholar |
Singh SP, Nodari R, Gepts P (1991b) Genetic diversity in cultivated common bean: I. Allozymes. Crop Science 31, 19–23.
| Genetic diversity in cultivated common bean: I. Allozymes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXhvVahsLo%3D&md5=a30eae4de75e68219b581fc282a68826CAS |
Smith B (1995) ‘The emergence of agriculture.’ (Scientific American Library: New York)
Sonnante G, Stockton T, Nodari RO, Becerra Velásquez VL, Gepts P (1994) Evolution of genetic diversity during the domestication of common-bean (Phaseolus vulgaris L.). Theoretical and Applied Genetics 89, 629–635.
| Evolution of genetic diversity during the domestication of common-bean (Phaseolus vulgaris L.).Crossref | GoogleScholarGoogle Scholar |
Staden R (1996) The Staden sequence analysis package. Molecular Biotechnology 5, 233–241.
| The Staden sequence analysis package.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xlt1Sgsb8%3D&md5=16557e841e64900c520389b437a4ddcaCAS |
Stewart CN, Halfhill MD, Warwick SI (2003) Transgene introgression from genetically modified crops to their wild relatives. Nature Reviews. Genetics 4, 806–817.
| Transgene introgression from genetically modified crops to their wild relatives.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXot1aksLc%3D&md5=ceffa605581b347139417da695dd3b9bCAS |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066.
| Seed banks and molecular maps: unlocking genetic potential from the wild.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXlsFSisrw%3D&md5=f13f988337a844632fca565639492c2bCAS |
Tenaillon MI, U’Ren J, Tenaillon O, Gaut BS (2004) Selection versus demography: a multilocus investigation of the domestication process in maize. Molecular Biology and Evolution 21, 1214–1225.
| Selection versus demography: a multilocus investigation of the domestication process in maize.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXltlKlsrk%3D&md5=ec65c77df64b33221b07715a6cf98c95CAS |
Tohme J, Gonzalez D, Beebe S, Duque MC (1996) AFLP analysis of gene pools of a wild bean core collection. Crop Science 36, 1375–1384.
| AFLP analysis of gene pools of a wild bean core collection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XmsFCkurk%3D&md5=45135bab45050255eb5a1361cfc3998cCAS |
Vasemagi A, Nilsson J, Primmer CR (2005) Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic salmon (Salmo salar L.). Molecular Biology and Evolution 22, 1067–1076.
| Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic salmon (Salmo salar L.).Crossref | GoogleScholarGoogle Scholar |
Wright SI, Gaut BS (2005) Molecular population genetics and the search for adaptive evolution in plants. Molecular Biology and Evolution 22, 506–519.
| Molecular population genetics and the search for adaptive evolution in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhs1Krurs%3D&md5=a2e0db2739c2269f97e257183c685259CAS |
Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS (2005) The effects of artificial selection on the maize genome. Science 308, 1310–1314.
| The effects of artificial selection on the maize genome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXks1Ciur0%3D&md5=9ddead049a4f0c302c398552ec2dffb8CAS |
Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M (2007) An Arabidopsis example of association mapping in structured samples. PLOS Genetics 3, e4
| An Arabidopsis example of association mapping in structured samples.Crossref | GoogleScholarGoogle Scholar |
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003) Single-nucleotide polymorphisms in soybean. Genetics 163, 1123–1134.
Zhu Q, Zheng X, Luo J, Gaut BS, Ge S (2007) Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Molecular Biology and Evolution 24, 875–888.
| Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXjtlersLo%3D&md5=dad43b88b3aa44d9e9824981e1743199CAS |