Genome-level identification of cell wall invertase genes in wheat for the study of drought tolerance
Hollie Webster A , Gabriel Keeble B , Bernard Dell A , John Fosu-Nyarko C , Y. Mukai D , Paula Moolhuijzen B , Matthew Bellgard B , Jizeng Jia E , Xiuying Kong E , Catherine Feuillet F , Frédéric Choulet F , International Wheat Genome Sequencing Consortium G and Rudi Appels B HA School of Biological Sciences and Biotechnology, Murdoch University, Perth, WA, Australia.
B Centre for Comparative Genomics, Murdoch University, Perth, WA, Australia.
C State Agricultural Biotechnology Centre, Murdoch University, Perth, WA, Australia.
D Laboratory of Plant Molecular Genetics, Division of Natural Science, Osaka Kyoiku University, Kashiwara, Osaka 582-8582, Japan.
E Key Laboratory of Crop Gene Resources and Germplasm Enhancement, Ministry of Agriculture, National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China.
F INRA UMR 1095, Génétique Diversité et Ecophysiologie des Céréales, 63100 Clermont-Ferrand, France.
G International Wheat Genome Sequencing Consortium, Executive Director Kellye Eversole. Email: eversole@eversoleassociates.com
H Corresponding author. Email: rappels@ccg.murdoch.edu.au
Functional Plant Biology 39(7) 569-579 https://doi.org/10.1071/FP12083
Submitted: 6 April 2012 Accepted: 17 May 2012 Published: 12 July 2012
Abstract
In wheat (Triticum aestivum L.) drought-induced pollen sterility is a major contributor to grain yield loss and is caused by the downregulation of the cell wall invertase gene IVR1. The IVR1 gene catalyses the irreversible hydrolysis of sucrose to glucose and fructose, the essential energy substrates which support pollen development. Downregulation of IVR1 in response to drought is isoform specific and shows variation in temporal and tissue-specific expression. IVR1 is now prompting interest as a candidate gene for molecular marker development to screen wheat germplasm for improved drought tolerance. The aim of this study was to define the family of IVR1 genes to enable: (1) individual isoforms to be assayed in gene expression studies; and (2) greater accuracy in IVR1 mapping to the wheat genetic map and drought tolerance QTL analysis. Using a cell wall invertase-specific motif as a probe, wheat genomics platforms were screened for the presence of unidentified IVR1 isoforms. Wheat genomics platforms screened included the IWGSC wheat survey sequence, the wheat D genome donor sequence from Aegilops tauschii Coss, and the CCG wheat chromosome 3B assembly: contig506. Chromosome-specific sequences homologous to the query motif were isolated and characterised. Sequence annotation results showed five previously unidentified IVR1 isoforms exist on multiple chromosome arms and on all three genomes (A, B and D): IVR1–3A, IVR1–4A, IVR1–5B, IVR1.2–3B and IVR1-5D. Including three previously characterised IVR1 isoforms (IVR1.1–1A, IVR1.2–1A and IVR1.1–3B), the total number of isoform gene family members is eight. The IVR1 isoforms contain two motifs common to cell wall invertase (NDPN and WECPDF) and a high degree of conservation in exon 4, suggesting conservation of functionality. Sequence divergence at a primary structure level in other regions of the gene was evident amongst the isoforms, which likely contributes to variation in gene regulation and expression in response to water deficit within this subfamily of IVR1 isoforms in wheat.
Additional keywords: gene family, grain yield, pollen sterility, sequencing, sucrose supply, water stress.
Introduction
In wheat (Triticum aestivum L.), grain number per ear is the main determinant of yield and is significantly reduced by pollen sterility when drought is incurred during pollen meiosis. Expression of the cell wall invertase (CW-INV) gene IVR1 in tapetal tissues that surround pollen during its formation is essential in maintaining pollen fertility. IVR1 functions to catalyse the irreversible hydrolysis of sucrose to glucose and fructose, the essential energy substrates that support pollen development. IVR1 is downregulated by water stress, which leads to pollen being starved of its energy source, resulting in sterility, reduced grain number and ultimately reduced yield (Koonjul et al. 2005; Parish et al. 2012; Powell et al. 2012).
In view of this evidence and the increasing frequency of drought events in Australia over the last decade, there has been growing demand for wheat pre-breeding research to elucidate the molecular determinants of IVR1 gene regulation and expression under limited and non-limited water conditions (Ji et al. 2010; Powell et al. 2012). Underpinning the interest for improved understanding of sugar metabolism during head development is the objective of breeding wheat cultivars capable of maintaining pollen fertility and thus grain set under drought conditions. Achieving this objective would better enable the genetic potential of existing wheat germplasm, capable of high yields in non-limited water environments, to be fully exploited in regions were water is frequently limited during the period in the growing season that coincides with head development and grain fill (Reynolds et al. 2005; Ji et al. 2010).
Evidence supporting molecular approaches to breeding wheat for stable pollen fertility was seen in recent studies by Ji et al. (2010), where wheat varietal-specific expression of IVR1 under water deficit conditions was examined. Results from this study were consistent with similar studies by Koonjul et al. (2005) and demonstrated a positive correlation between IVR1 expression and pollen fertility. However, the degree of IVR1 downregulation in response to water deficit showed significant differences between varieties, in that certain varieties maintained gene expression levels under water stress, whereas others did not. This result is consistent with IVR1 being a strong candidate gene for drought tolerance quantitative trait loci (QTL) analysis (Ji et al. 2010).
Plant invertases (β-D-fructofuranosidase EC 3.2.1.26) constitute a family of enzymes that hydrolyse sucrose to glucose and fructose. In plants, three types of invertases have been identified: cell wall (CW-INV), vacuolar (V-INV) and cytoplasmic (C-INV). Invertases have roles in several plant physiological processes related to long-distance nutrient allocation as well as regulating developmental processes, hormone responses and biotic and abiotic interactions (Lalonde et al. 1997; Tymowska-Lalanne and Kreis 1998; Roitsch and Gonzalez 2004; Ruan et al. 2010). In order to provide a source of energy, sucrose is transported from leaves to the stem, reproductive structures and roots (Ruan et al. 2010). The sucrose molecule is cleaved at the α1-β2-glycosidic bond, either by invertases or sucrose synthase (SST). Invertases catalyse the irreversible hydrolysis of sucrose to glucose and fructose and SST catalyses reversible cleavage of sucrose to UDPglucose and fructose (Roitsch and Gonzalez 2004). Upon cleavage of sucrose, glucose and fructose are transported into the cells via hexose transporters located in the plasma membrane (Bush 1999).
The invertase gene family consists of many members within each of the three broad functional groups (i.e. CW-INV, V-INV and C-INV) (Tymowska-Lalanne and Kreis 1998). In the present study, the gene nomenclature used defines IVR1 isoforms as belonging to the cell wall invertase (CW-INV) gene subfamily in wheat. The term isoform is used to designate different proteins derived from related genes. A widely accepted explanation of multiple invertase isoform existence within each of the functional groups is that this allows for greater flexibility in the control of sucrose metabolism, translocation and storage under different internal and external stimuli and at various developmental stages or tissues, or at the whole-plant level (Tymowska-Lalanne and Kreis 1998; Akhunov et al. 2003). Koonjul et al. (2005) found that individual IVR1 and V-INV isoforms were expressed in a highly specific manner in response to water availability, with respect to both tissue type and developmental stage. Similar results were recently reported by Crismani et al. (2011), who showed that in rice, closely related IVR1 isoforms within anther-specific transcripts exhibited clear temporal variation under non-limiting water conditions.
Studies of invertase transcription regulation conducted in several different crop species have found that transcription is influenced by a range of signals including; low oxygen, plant hormones, sugars and environmental and pathogenic cues (Long et al. 2002; Koch 2004; Proels and Roitsch 2009). In studies specifically examining the role of sucrose in regulating CW-INV expression, varied results were reported; some CW-INV appear to have expression reduced by sucrose accumulation, whereas others isoforms are upregulated (Kaufman et al. 1973; Sturn and Chrispeels 1990). Tymowska-Lalanne and Kreis (1998) suggest that variable CW-INV transcription under the control of sucrose signalling may be related to different isoforms being regulated in a highly specific manner, as a reflection of subtle but important differences in their function.
The aim of this paper was to define IVR1 isoforms at the genome level in wheat, in order to provide a basis for greater accuracy in discriminating between the isoforms that are expressed in developing wheat heads under normal and water deficit conditions. Evidence from this study suggests that characterisation of the IVR1 isoforms is essential to further gene expression studies as it will enable definitive assays of single isoforms. IVR1 isoform-specific assays will permit variation in spatial and temporal expression to be analysed in the context of improving understanding of IVR1 gene regulation and functionality.
Materials and methods
Wheat BAC sequencing
The assembly of BACs from flow-sorted chromosome 3B DNA was as described by Paux et al. (2008) and utilised a modified HICF. SNaPshot protocol was in summary, five restriction endonucleases were used to digest DNA samples. Anchoring of the BAC contigs to a genetic map was conducted using established SSR markers as well as probes for insertion-based polymorphisms, using a mapping population derived from a cross between wheat varieties Chinese Spring × Renan and genetic lines carrying chromosomal deletions (Paux et al. 2008). For contig506 (ctg506), the minimum tiling path was established using the FPC software at a high stringency (1e–75) and refined by analyses using LTC software (Frenkel et al. 2010). Sequencing was conducted at 5–8× coverage and the Solexa-Illumina sequencing was conducted at >300× coverage using 75 base pair reads. Additional 454 sequencing using 8-kb mate-pair libraries resolved ambiguities in the sequence assembly. The 480-kb portion of ctg506 (1.3 Mb) analysed in this paper has been submitted to GenBank (GenBank submission pending).
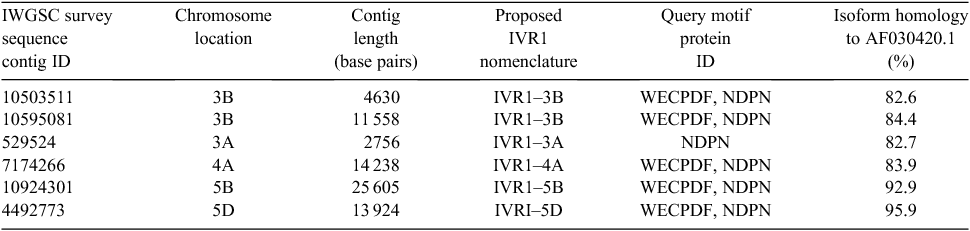
Identification of IVR1 isoforms and sequence annotation
Access to the genome survey sequences for the chromosome arms of wheat before publication (http://urgi.versailles.inra.fr/Species/Wheat/Sequence-Repository, accessed 10 May 2012) was provided by the International Wheat Genome Sequencing Consortium (IWGSC). To screen for the presence of previously unidentified IVR1 isoforms a BLASTN query was conducted across the entire wheat survey sequence using two invertase-specific motifs: (1) 5′CAGATCCAAATGGTACGTA′3, residues which span a β-fructosidase motif (NDPN) that is common to all invertase genes (i.e. CW-INV, V-INV, C-INV) and (2) 5′TGGGAATGCCCCGATTTC′3, residues which span a catalytic domain (WECPDF) that is present only in CW-INV isoforms (Tymowska-Lalanne and Kreis 1998; Koonjul et al. 2005). Determination of sequence-based IVR1 categorisation required the presence of both the NDPN and WECPDF motifs. The BLASTN analysis was conducted across all available chromosome arm datasets and yielded subject sequence contigs identified by accession numbers, which were then used to obtain the respective contigs from the specific chromosome arm on which they were located (Table 1). The contig sequences were imported into Geneious Pro (ver. 5.5.6, http://www.genious/com/) for alignment and annotation using the IVR1 full-length cDNA sequence, accession AF030420.1, as a reference.
Plant material and DNA extraction
Wheat (Triticum aestivum L. cv. Westonia and cv. Kauz) seed stocks were provided by the Department of Agriculture and Food Western Australia (DAFWA). Seed stocks were pre-germinated at 4°C for 14 days and then planted into free draining pots with a uniformly prepared soil medium consisting of two parts loam, one part river sand and 3 g of basal fertiliser (mg kg–1: 1220 N, 368 P, 819 K) and grown in controlled glasshouse conditions with ambient temperature during the plant growth period of 9°C (night) and 18°C (day). Pots were watered three times daily using an irrigation mat system to maintain average soil volumetric water content of 60%. Leaf tissue samples were taken for DNA extraction at the early tillering phase (Zadok’s growth scale stage Z20), frozen in liquid nitrogen and DNA extraction was performed using a generic phenol-chloroform extraction method. The extracted DNA was quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher, Scoresby, Vic., Australia).
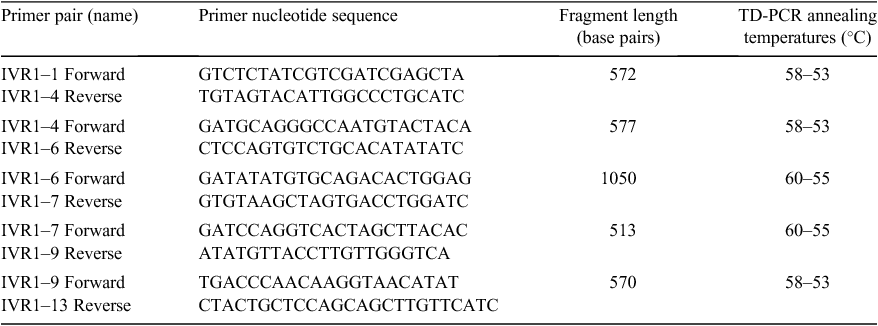
IVR1 sequencing and analysis
The genomic sequence ctg506 (GenBank submission pending), as defined in this manuscript, was located on chromosome 3B of Chinese Spring and contained IVR1.1–3B. This isoform was used as the reference sequence to design primers (each spanning intron–exon junctions) to amplify a total of five overlapping fragments spanning the entire length of IVR1.1–3B (Table 2). Primers were designed with Primer3 ver. 0.4.0 (http://primer3.sourceforge.net/, accessed 5 March 2011). Each PCR reaction was 20 µl, containing: 200 ng of template DNA, 10 pmol each of a forward and reverse primer pair (specific to an individual genomic fragment), 200 µM dNTP, 2.5 mM MgCl2, 1× reaction buffer, 6 µl of 1% cresol red and 2.5 units of Taq polymerase (Fisher Biotech, Perth, WA, Australia). Touch down PCR (TD-PCR) conditions were: incubation for 3 min at 94°C, followed by 35 cycles of 1 min denaturation (94°C), 30 s annealing (decreasing by 1°C per cycle until the primer-specific annealing temperature was reached, as in Table 2) and 30 s extension (72°C). An additional 10 minutes extension at 72°C was also performed.
|
The TD-PCR products were separated on a 2% agarose gel and DNA purified using a Wizard SV Gel and PCR clean up system (Promega Corp., Annandale, NSW, Australia). The purified genomic DNA fragments were cloned by ligation to the pGEM-T Easy cloning vector (Promega Corp.). Both strands of the cloned IVR1 isoform fragments were sequenced with the Sp6 and T7 promoter using BigDye3.1 (BDV3.1 plus dgtp) termination chemistry and were run on an ABI Prism XL 3730 sequencer (Applied Biosystems, Foster City, CA, USA).
Results
The genome level structure of wheat IVR1 isoforms on chromosome 3B
Our sequence assembly of a BAC contig from chromosome 3B (ctg506) provided the genome sequence of two IVR1 isoforms, IVR1.1–3B and IVR1.2–3B, in addition to the two previously characterised genes on chromosome 1A (Francki et al. 2006).
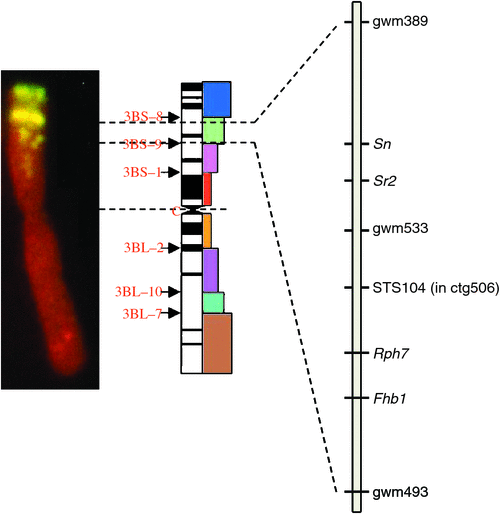
The ctg506 is between the Sr2 rust resistance and fusarium head blight resistance (Fhb1) loci and includes the molecular marker STS104 (genetic map in Fig. 1). The assembly initially comprised 12 BAC clones, with estimated insert sizes of 70–180 kb, compiled into a minimum tiling path. The linear sequence was established using a combination of Sanger and Solexa-Illumina sequencing technologies. The initial assembly had 21 contigs. Subsequent analyses of the FPC based assemblies and additional 454 sequencing of 8-kb mate pair libraries refined the assembly.
|
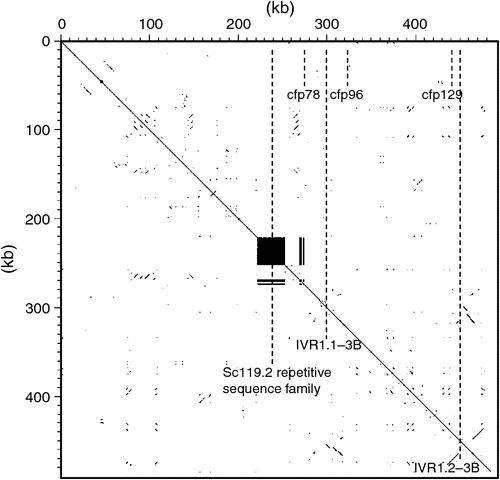
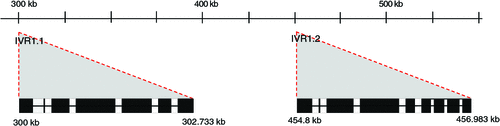
Figure 2 shows the details for 480 kb of ctg506 to indicate the overall structure of this genome sequence and the location of the IVR1 isoforms. The highly repetitive region upstream from IVR1.1–3B comprises members of the Sc119.2 repetitive DNA family that are widely used as chromosome markers (left-hand panel Fig. 1; McIntyre et al. 1990) and are distributed in the ctg506 region of chromosome 3B at a relatively low representation compared with other sites in the genome (note the more intense band slightly distal to the region of interest). A SNP (BF293133) was found further downstream on the ctg506 genome segment shown in Fig. 2 and has been assigned a genetic location on chromosome 3B in the position expected for ctg506 (Zhang et al. 2008a), providing additional confirmation of the chromosomal location of the sequences analysed.
|
Annotation of the 480-kb genome sequence was conducted using GenScan (http://genes.mit.edu/GENSCAN.html, 9 May 2012) as well as Augustus and EuGene (in TriAnnot http://urgi.versailles.inra.fr/Species/Wheat/Triannot-Pipeline/, accessed 9 May 2012, Leroy 2012). The results consistently showed two IVR1 isoforms at coordinates 300 kb and 454 kb in the genome sequence. In Fig. 3, the primary structure for the IVR1 isoforms is presented based on manual refinement of the annotations in the Geneious program, using AF030420.1 as a reference sequence to identify intron-exon boundaries. The IVR1.1–3B isoform has seven exons and is clearly closely related to the reference cell wall invertase AF030420.1, including a characteristic small exon of three amino acids (DPN, part of the NDPN motif) and a conserved catalytic domain in exon 4 (WECPDF). The IVR1.2–3B isoform also has the DPN and WECPDF motifs but has a nine exon structure and greater sequence divergence in other parts of the coding regions (as compared with the reference AF030420.1), particularly in exon 3.
|
The IVR1 gene family in wheat
Sequence analysis of the cloned genomic DNA TD-PCR products amplified from exon 3 and exon 7, in both Westonia and Kauz, provided the initial evidence for the existence of additional IVR1 isoforms in the wheat genome that had not yet been identified. The primers used to assay the fragments in question spanned exon-intron boundaries and thus were expected to have high specificity to assay IVR1.1–3B, given they were designed using the Chinese Spring chromosome 3B sequence. Three homologous groupings were apparent in both the Westonia and Kauz sequences of exons 3 and 7, one group of which was homologous to IVR1.1–3B (ctg506) but the remaining two did not show homology to any of the available annotated IVR1 sequences for wheat, including: IVR1.1–3B (ctg506), IVR1.1–1A (GenBank submission pending), IVR1.2–1A (GenBank submission pending) and the full length IVR1 cDNA AF030420.1. This evidence suggested at least two additional IVR1 isoforms existed and that there was likely to be an IVR1 subfamily in wheat.
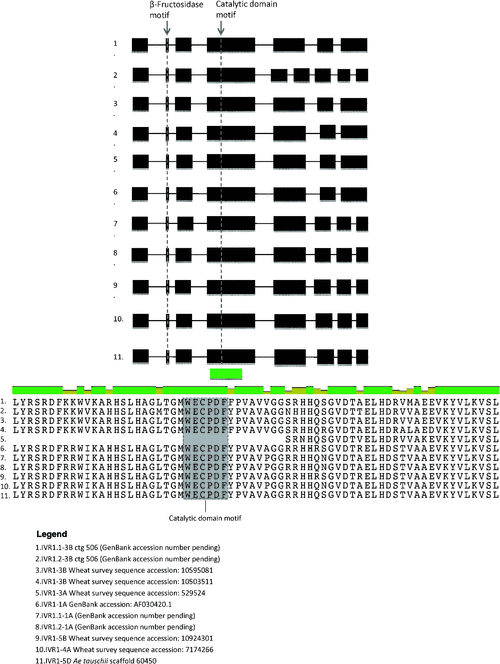
The IWGSC survey sequences for the chromosome arms of wheat (http://urgi.versailles.inra.fr/Species/Wheat/) enabled four previously unidentified isoforms of the IVR1 subfamily to be identified using the invertase-specific motif NDPN and the CW-INV-specific motif WECPDF, as probes. Using these probes, to screen the wheat D genome donor sequence from Triticum tauschii (J. Jia, unpubl. data) and the ctg506 chromosome 3B assembly, two additional uncharacterised IVR1 isoforms were also identified. Figure 4 shows the isoform gene structure determined using the full-length IVR1 cDNA AF030420.1 as a reference. The characterised and annotated IVR1 isoforms show the conserved motifs DPN (exon 2) and WECPDF (exon 4). Conservation around the WECPDF motif spans the entire length of exon 4 (see protein sequence of exon 4 in the lower panel of Fig. 4). Divergence in intron-exon structure is apparent, with five members for the IVR1 family having eight exons (IVR1.1–1A, IVR1.2–1A, IVR1–4A, IVR1–5B, IVR1-5D), two members having seven exons (IVR1.1–3B, IVR1–3A) and IVR1.2–3B comprising nine exons.
|
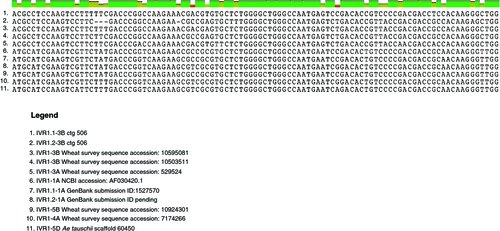
The evidence presented in Fig. 4 was based on the screening of individual chromosome arm libraries, as distinct from counting sequence variants. These data suggest that at least eight IVR1 isoforms exist in wheat, including: IVR1.1–1A, IVR1.2–1A, IVR1–3A, IVR1.1–3B, IVR1.2–3B, IVR1–4A, IVR1–5B and IVR1-5D. From these results we propose that using their chromosomal locations is a useful way to identify the isoforms. In Fig. 5 the nucleotide sequence of exon 4 (containing the WECPDF motif) is shown and illustrates that IVR1 isoforms are identifiable by their distributions of SNPs, enabling clear differentiation between the isoforms. We note that defined segments of IVR1.1–1A and IVR1.2–1A were assigned to 1A using genetic stocks (Francki et al. 2006), but our survey sequence analysis did not support this chromosomal location. Thus, it is possible that fewer IVR1 genes exist and that IVR1.1–1A is actually IVR1–4A and IVR1.2–1A is IVR1–5B. This needs further confirmation.
|
Screening of the IWGSC survey sequence, the ctg506 chromosome 3B assembly and the D genome sequence with PCR primers previously used to study the expression of IVR1 in wheat (Koonjul et al. 2005; Ji et al. 2010), indicated that several IVR1 isoforms on different chromosomes were assayed by these primers. As shown in Fig. 6a, the primers by Ji et al. (2010) are predicted to assay IVR1–1A, IVR1–4A and IVR1–5B. The primers used by Koonjul et al. (2005) would similarly assay IVR1.1–3B and IVR1–5B (Fig. 6b).

|
Comparative genomics of cell wall invertases in wheat and rice
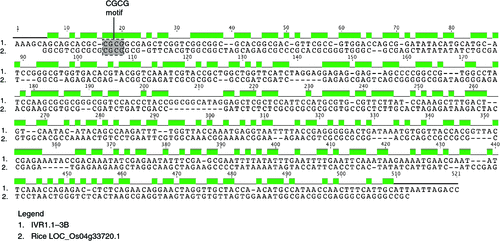
Within the rice genome sequence eight cell wall invertase isoforms has been identified (Cho et al. 2005; Sturm and Tang 1999). The extensive phenotypic analyses associated with genes and regions of rice provide a reference for studies in wheat. Phylogenetic analysis indicated that among the cell wall invertase isoforms in rice, the LOC_Os04g33720 sequence on chromosome 4 was most closely related to the AF030420.1 sequence. The LOC_Os04g33720 gene is preferentially transcribed in anther tissues although there is expression at lower levels (i.e. 30% or less) in other tissues (Sharma et al. 2012). It was of interest to align the 5′ upstream regions from the LOC_Os04 g33720 gene of rice (–507 to 0) with available upstream regions from the wheat IVR1 isoforms, IVR1.1–3B (–1069 to –562) and IVR1-5D (–1777 to –1356), to identify conserved motifs.
The analysis shown in Fig. 7 provides evidence that motifs conserved between rice and wheat are located upstream of IVR1.1–3B, indicating that these may be significant for anther-specific expression of IVR1 isoforms. Similar results were obtained for IVR1.2–3B and IVR1-5D upstream regions aligned with the respective rice region.
|
Discussion
The data in the present paper have further defined the IVR1 subfamily in wheat at the genome level. Complexity at this level was suggested by Koonjul et al. (2005) and the genome-level analysis of IVR1 has indicated that the primers used to assay transcripts in drought experiments (Koonjul et al. 2005; Ji et al. 2010) actually assay several isoforms from different chromosomal locations. The significance of these findings is that transcription studies need to define the chromosomal location of the isoform being assayed so that suitable molecular markers can be transferred to wheat breeding.
As in cotton (Udall et al. 2007; Rapp et al. 2010), wheat shows homeologous gene silencing (Mochida et al. 2004; Bottley and Koebner 2008; Zhang et al. 2008b). In an analysis of a panel of 16 wheat varieties, Bottley and Koebner (2008) identified homoeolog non-expression for 15 single-copy genes. The detailed expression profiles of eight genes indicated that only two varieties of the 16 shared the same pattern of silencing and thus, the homoeologous silencing observed in wheat is likely to be significant in generating phenotypic variation. In this context it is evident that in relating variation in expression of IVR1 isoforms in wheat to drought tolerance it is important to define the chromosomal origin of the IVR isoform being studied in transcriptional analyses.
There is growing evidence to suggest that sucrose conversion by cell wall invertases in the apoplastic space is one of the key rate limiting steps in carbohydrate metabolism in reproductive structures and supplying photosynthetic assimilates for storage (Eschrich 1980). The basis of this dependency is that developing pollen microspores are symplastically isolated and thus, must receive carbohydrates via apoplastic transport in which IVR1 isoforms play a critical role (Roitsch 1999; Sturm and Tang 1999). Impeded apoplastic transport can be attributed to the downregulation of IVR1 in anthers, which results in pollen being starved of the energy substrate, as there is no alternate pathway for the delivery of sucrose. The breakthrough in establishing the central role of CW-INV’s in phloem uploading of sugars was in the study of the Nin88 CW-INV gene in tobacco (Goetz et al. 2001), which was specifically expressed in the tapetum and pollen. In their study, Goetz et al. (2001) demonstrated that tissue-specific suppression of the Nin-88 gene, under the control of its own promoter sequence, resulted in defects in the early stages of pollen development and thus sterile pollen.
In studies examining the selective downregulation of IVR1 isoforms under water deficit conditions in wheat, Koonjul et al. (2005) reported that gene expression was highly specific to individual isoforms both spatially and temporally. Similar conclusions were drawn by Godt and Roitsch (1997) and Sturm and Tang (1999). In the Koonjul et al. (2005) study, water deficit applied specifically during the period just before and following pollen meiosis showed immediate and significant downregulation of one IVR1 isoform (IVR1S) but no decline in another (IVR1 L). Conversely, in the control plants (where watering was maintained) the IVR1S isoform also declined over the duration of pollen development whereas IVR1 L increased. According to the results shown in Fig. 6b, which highlighs the PCR products assayed by the Koonjul et al. (2005) IVR1 primers, we propose that the two IVR1 isoforms assayed in this gene expression analysis were located on chromosome 3 (IVR1.1–3B) and chromosome 5 (IVR1–5B). However, determination of which isoform was located on chromosomes 3 and 5 was not possible without access to isoform-specific PCR product sequences.
The Koonjul et al. (2005) invertase gene expression analysis also highlighted that two genes (IVR3 and IVR5), closely related to the IVR1 family, did not show decline in expression under drought stress. The IVR3 and IVR5 genes were reported as being members of the CW-INV and V-INV families, respectively. In the work related to the current study, detailed examination of the IVR3 (accession AF030421) and IVR5 (accession AF069309) gene sequences confirmed that IVR5 belongs to the V-INV family but that IVR3 is not a CW-INV or a V-INV isoform. Our results show that the IVR3 sequence is: (i) highly divergent from the IVR1 family both at a primary sequence level and exon structure; (ii) located on chromosome 6A in wheat (IWGSC Survey Sequence contig-4429358); and (iii) a member of the fructan 1-exohydrolase gene family, with its closest relative being the 1-FEHw1 (accession AJ516025) gene in wheat.
The studies in the present paper provide greater resolution in defining IVR1 isoforms for expression studies. The comparison in Fig. 7 showed that the 5′-upstream region for IVR1.1–3B (ctg506: 298 676 kb to 300 000 kb) had significant similarity to an equivalent region for LOC_Os04 g33720 (ch4: 20 411 920 kb to 20 412 390 kb). The shared motifs included the CGCG box and CGACG motif. The CGCG box has been characterised in Arabidopsis (Yang and Poovaiah 2002) and often occurs as multiple elements as is the case for region shown in Fig. 7. The CGACG motif has been studied in the cis-control of amylase (Amy3D) expression in rice (Hwang et al. 1998). Within the IVR1.1–3B region the ACTCAT (related to osmoregulation), AGAAA (within a pollen-specific promoter) and CATGTG (related to drought response) motifs can be identified (ProFITS site: http://bioinfo.cau.edu.cn/ProFITS/, accessed 27 April 2012) and their presence is consistent with IVR1 expression being significant in maintaining a supply of hexose sugars to the developing pollen. We note that IVR1.1–3B is located in the region of chromosome 3BS where a QTL for variation in early head development, a stage that is particularly sensitive to the effects of water stress (Maccaferri et al. 2008).
Gene duplication is widely recognised as a platform from which gene families can evolve through mechanisms of chromosomal remodelling, locus deletion or point mutation (Wang et al. 2010). The data in this manuscript further support this hypothesis, with regards to the evolution of complex gene families in wheat (derived from a common ancestral gene), by providing evidence of the existence of a multi member IVR1 subfamily, which has a shared functionality in hydrolysing hexoses in reproductive structures. We propose the sequence divergence that exists amongst the eight IVR1 isoforms, located on four separate chromosome arms of the wheat genome, is likely to have causal links to subtle differences in isoform functionality and regulatory mechanisms, particularly in response to changing water availability conditions. It is plausible to suggest that sequence variation in the IVR1 subfamily provides wheat the flexibility to maintain carbohydrate supply to its developing reproductive structures, thus ensuring viable pollen development even in drought conditions. Gene expression studies remain the foundation methodology by which the relationship between the sequence variation of the IVR1 isoforms and their corresponding functional differences can be better understood. The identification of the chromosomal origins of the IVR1 isoforms undertaken in this study will now enable more accurate transcriptional analysis and improved capacity to relate variation in IVR1 isoform expression to drought tolerance in wheat.
Acknowledgements
The authors wish to thank Dr Philippe Leroy for his help in annotation of the ctg506 wheat genome sequence.
References
Akhunov ED, Goodyear AW, Geng S, Qi L, Echalier B, Gill BS, Miftahudin , Gustafson JP, Lazo G, Chao S, Anderson OD, Linkiewicz AM, Dubcovsky J, La Rota M, Sorrells ME, Zhang D, Nguyen HT, Kalavacharla V, Hossain K, Kianian SF, Peng J, Lapitan NL, Gonzalez-Hernandez JL, Anderson JA, Choi DW, Close TJ, Dilbirligi M, Gill KS, Walker-Simmons K, Steber C, McGuire PE, Qualset CO, Dvorak J (2003) The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Research 13, 753–763.| The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjs1Oqtbw%3D&md5=97ef3a98ae9d11a54808ffe706901603CAS |
Bottley A, Koebner RM (2008) Variation for homoeologous gene silencing in hexaploid wheat. The Plant Journal 56, 297–302.
| Variation for homoeologous gene silencing in hexaploid wheat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtlChu7rL&md5=b08073d67b646ce29d03e0e9ec55388aCAS |
Bush DR (1999) Sugar transporters in plant biology. Current Opinion in Plant Biology 2, 187–191.
| Sugar transporters in plant biology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXkt1Cgs70%3D&md5=9c0f5c2dfc40ea86a91b82d0d9b12d05CAS |
Cho JI, Lee SK, Ko S, Kim HK, Jun SH, Lee HY, Bhoo SH, Lee KW, An G, Hahn TR, Jeon JS (2005) Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.). Plant Cell Reports 24, 225–236.
| Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXlt1Wmtbk%3D&md5=a10d34148bad5d131949b0d04ac61ef2CAS |
Crismani W, Kapoor S, Able JA (2011) Comparative transcriptomics reveals 129 transcripts that are temporally regulated during anther development and meiotic progression in both bread wheat (Triticum aestivum) and rice (Oryza sativa). International Journal of Plant Genomics 2011, 931 898
| Comparative transcriptomics reveals 129 transcripts that are temporally regulated during anther development and meiotic progression in both bread wheat (Triticum aestivum) and rice (Oryza sativa).Crossref | GoogleScholarGoogle Scholar |
Eschrich W (1980) Free space invertase, its possible role in phloem unloading. Berichte Der Deutschen Botanischen Gesllschaft 93, 363–378.
Francki MG, Walker E, Forster JW, Spangenberg G, Appels R (2006) Fructosyltransferase and invertase genes evolved by gene duplication and rearrangements: rice, perennial ryegrass, and wheat gene families. Genome 49, 1081–1091.
| Fructosyltransferase and invertase genes evolved by gene duplication and rearrangements: rice, perennial ryegrass, and wheat gene families.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXlslGhsw%3D%3D&md5=b860cfa492e4c8779dc17de62d402491CAS |
Frenkel Z, Paux E, Mester D, Feuillet C, Korol A (2010) LTC: a novel algorithm to improve the efficiency of contig assembly for physical mapping in complex genomes. BMC Bioinformatics 11, 584
| LTC: a novel algorithm to improve the efficiency of contig assembly for physical mapping in complex genomes.Crossref | GoogleScholarGoogle Scholar |
Godt DE, Roitsch T (1997) Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiology 115, 273–282.
| Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXmt1ynur0%3D&md5=b5747dcb8732371c43d82d272a1468c1CAS |
Goetz M, Godt DE, Guivarc’h A, Kahmann U, Chriqui D, Roitsch T (2001) Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proceedings of the National Academy of Sciences of the United States of America 98, 6522–6527.
| Induction of male sterility in plants by metabolic engineering of the carbohydrate supply.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXktVWktLc%3D&md5=8c3e4c1e6a720a19e3d1052d1c3e08a7CAS |
Hwang YS, Karrer EE, Thomas BR, Chen L, Rodriguez RL (1998) Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Molecular Biology 36, 331–341.
| Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXht1Wht7w%3D&md5=7f78328c96568d14dba75a997e1469afCAS |
Ji X, Shiran B, Wan J, Lewis DC, Jenkins CLD, Condon AG, Richards RA, Dolferus R (2010) Importance of pre-anthesis anther sink strength for maintenance of grain number during reproductive stage water stress in wheat. Plant, Cell & Environment 33, 926–942.
| Importance of pre-anthesis anther sink strength for maintenance of grain number during reproductive stage water stress in wheat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXnvVagsb4%3D&md5=0bde21dd472066df476747943a563387CAS |
Kaufman PB, Ghosheh NS, Lacroix JD, Soni SL, Ikuma H (1973) Regulation of invertase levels in Avena stem segments by gibberellic acid, sucrose, glucose, and fructose. Plant Physiology 52, 221–228.
| Regulation of invertase levels in Avena stem segments by gibberellic acid, sucrose, glucose, and fructose.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE3sXlt1ahtrg%3D&md5=c3a5879a520b8b9e2a33a2b2d18c8fc2CAS |
Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinion in Plant Biology 7, 235–246.
| Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjvVams7s%3D&md5=00a5f4a3d445a5533afac7c4f91e2127CAS |
Koonjul PK, Minhas JS, Nunes C, Sheoran IS, Saini HS (2005) Selective transcriptional down-regulation of anther invertases precedes the failure of pollen development in water-stressed wheat. Journal of Experimental Botany 56, 179–190.
Lalonde S, Beebe DU, Saini HS (1997) Early signs of disruption of wheat anther development associated with the induction of male sterility by meiotic-stage water deficit. Sexual Plant Reproduction 10, 40–48.
| Early signs of disruption of wheat anther development associated with the induction of male sterility by meiotic-stage water deficit.Crossref | GoogleScholarGoogle Scholar |
Leroy PI, Guilhot N, Sakai H, Bernard A, Choulet F, Theil S, Reboux S, Amano N, Fluetre T, Pelegrin C, Ohyanagi H, Seidel M, Giacomoni F, Reichstadt M, Alaux M, Gicquello E, Legeai F, Cerutti L, Numa H, Tanaka T, Mayer K, Itoh T, Quesneville H, Feuillet C (2012) TriAnnot: a versatile and high performance pipeline for the automated annotation of plant genomes. Frontiers in Plant Genetics and Genomics 3, 1–14.
| TriAnnot: a versatile and high performance pipeline for the automated annotation of plant genomes.Crossref | GoogleScholarGoogle Scholar |
Long JC, Zhao W, Rashotte AM, Muday GK, Huber SC (2002) Gravity-stimulated changes in auxin and invertase gene expression in maize pulvinal cells. Plant Physiology 128, 591–602.
| Gravity-stimulated changes in auxin and invertase gene expression in maize pulvinal cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XhsVSqsr0%3D&md5=0ba19bcda19379404efb0a211ad77f0eCAS |
Maccaferri M, Sanguineti MC, Corneti S, Ortega JL, Salem MB, Bort J, DeAmbrogio E, del Moral LF, Demontis A, El-Ahmed A, Maalouf F, Machlab H, Martos V, Moragues M, Motawaj J, Nachit M, Nserallah N, Ouabbou H, Royo C, Slama A, Tuberosa R (2008) Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics 178, 489–511.
| Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability.Crossref | GoogleScholarGoogle Scholar |
McIntyre CL, Pereira S, Moran LB, Appels R (1990) New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 33, 635–640.
| New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXktVaisrw%3D&md5=14d80f1e9f71155c17db34e653901562CAS |
Mochida K, Yamazaki Y, Ogihara Y (2004) Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Molecular Genetics and Genomics 270, 371–377.
| Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags.Crossref | GoogleScholarGoogle Scholar |
Parish RW, Phan HA, Iacuone S, Li SF (2012) Tapetal development and abiotic stress: a centre of vulnerability. Functional Plant Biology 39, 553–559.
| Tapetal development and abiotic stress: a centre of vulnerability.Crossref | GoogleScholarGoogle Scholar |
Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W, Lagudah E, Somers D, Kilian A, Alaux M, Vautrin S, Berges H, Eversole K, Appels R, Safar J, Simkova H, Dolezel J, Bernard M, Feuillet C (2008) A physical map of the 1-gigabase bread wheat chromosome 3B. Science 322, 101–104.
| A physical map of the 1-gigabase bread wheat chromosome 3B.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFyhurnK&md5=00d7f6d5127f308f3c4991c38cc9c4cfCAS |
Powell N, Ji X, Ravash R, Edlington J, Dolferus R (2012) Yield stability for cereals in a changing climate. Functional Plant Biology 39, 539–552.
| Yield stability for cereals in a changing climate.Crossref | GoogleScholarGoogle Scholar |
Proels RK, Roitsch T (2009) Extracellular invertase LIN6 of tomato: a pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by a diurnal rhythm. Journal of Experimental Botany 60, 1555–1567.
| Extracellular invertase LIN6 of tomato: a pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by a diurnal rhythm.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXltFWhsrs%3D&md5=14efdcd29c3a7ae2a1ee488e831c8a1aCAS |
Rapp RA, Haigler CH, Flagel L, Hovav RH, Udall JA, Wendel JF (2010) Gene expression in developing fibres of upland cotton (Gossypium hirsutum L.) was massively altered by domestication. BMC Biology 8, 139
| Gene expression in developing fibres of upland cotton (Gossypium hirsutum L.) was massively altered by domestication.Crossref | GoogleScholarGoogle Scholar |
Reynolds M, Pellegrineschi A, Slovmand B (2005) Sink-limitation to yield and biomass: a summary of some investigations in spring wheat. Annals of Applied Biology 146, 39–49.
| Sink-limitation to yield and biomass: a summary of some investigations in spring wheat.Crossref | GoogleScholarGoogle Scholar |
Roitsch T (1999) Source–sink regulation by sugar and stress. Current Opinion in Plant Biology 2, 198–206.
| Source–sink regulation by sugar and stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXkt1Cgs7s%3D&md5=a86f57c55c4dcc8bf210ba7509f3f336CAS |
Roitsch T, Gonzalez MC (2004) Function and regulation of plant invertases: sweet sensations. Trends in Plant Science 9, 606–613.
| Function and regulation of plant invertases: sweet sensations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVaksLvO&md5=a850db666203fb6c7184ca1f0d1200acCAS |
Ruan Y-L, Jin Y, Yang Y-J, Li G, Boyer JS (2010) Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Molecular Plant 3, 942–955.
| Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsFantLjL&md5=38438beb501abb116c83ee8ab3740feeCAS |
Sharma R, Agarwal P, Ray S, Deveshwar P, Sharma P, Sharma N, Nijhawan A, Jain M, Singh AK, Singh VP, Khurana JP, Tyagi AK, Kapoor S (2012) Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Functional & Integrative Genomics 12, 229–248.
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Functional & Integrative Genomics 4, 12–25.
| Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhsl2hsbc%3D&md5=b953a3bbd6edc9d18081ada89308934fCAS |
Sturm A, Tang GQ (1999) The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends in Plant Science 4, 401–407.
| The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning.Crossref | GoogleScholarGoogle Scholar |
Sturn A, Chrispeels MJ (1990) cDNA cloning of carrot extracellular beta-fructosidase and its expression in response to wounding and bacterial wounding. Plant Cell 2, 1107–1119.
Tymowska-Lalanne Z, Kreis M (1998) The plant invertases: physiology, biochemistry, and molecular biology. Advances in Botanical Research 28, 71–117.
| The plant invertases: physiology, biochemistry, and molecular biology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXhtlGgs7k%3D&md5=567ff418c61eb739a9f106a9914541d2CAS |
Udall JA, Flagel LE, Cheung F, Woodward AW, Hovav R, Rapp RA, Swanson JM, Lee JJ, Gingle AR, Nettleton D, Town CD, Chen ZJ, Wendel JF (2007) Spotted cotton oligonucleotide microarrays for gene expression analysis. BMC Genomics 8, 81
| Spotted cotton oligonucleotide microarrays for gene expression analysis.Crossref | GoogleScholarGoogle Scholar |
Wang E, Xu X, Zhang L, Zhang H, Lin L, Wang Q, Li Q, Ge S, Lu BR, Wang W, He Z (2010) Duplication and independent selection of cell-wall invertase genes GIF1 and OsCIN1 during rice evolution and domestication. BMC Evolutionary Biology 10, 108
| Duplication and independent selection of cell-wall invertase genes GIF1 and OsCIN1 during rice evolution and domestication.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXnvFCgu7g%3D&md5=f4b71578d50150c86ddf71b37d751826CAS |
Yang T, Poovaiah BW (2002) A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. Journal of Biological Chemistry 277, 45 049–45 058.
| A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xosl2murs%3D&md5=87b8d331ee12ad7472823ad0b38e4856CAS |
Zhang W, Chao S, Manthey F, Chicaiza O, Brevis JC, Echenique V, Dubcovsky J (2008a) QTL analysis of pasta quality using a composite microsatellite and SNP map of durum wheat. Theoretical and Applied Genetics 117, 1361–1377.
| QTL analysis of pasta quality using a composite microsatellite and SNP map of durum wheat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtlSgsLnI&md5=37ff1bbec3372d7b29e5baae0c3e65bdCAS |
Zhang J, Huang S, Fosu-Nyarko J, Dell B, McNeil M, Waters I, Moolhuijzen P, Conocono E, Appels R (2008b) The genome structure of the 1-FEH genes in wheat (Triticum aestivum L.): new markers to track stem carbohydrates and grain filling QTLs in breeding. Molecular Breeding 22, 339–351.
| The genome structure of the 1-FEH genes in wheat (Triticum aestivum L.): new markers to track stem carbohydrates and grain filling QTLs in breeding.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtVagurrM&md5=f03e7cf801b7300e3674ad2a3351a08aCAS |