The integration of activity in saline environments: problems and perspectives
John M. CheesemanA Department of Plant Biology, University of Illinois, Urbana, IL 61801, USA. Email: j-cheese@illinois.edu
This paper originates from a presentation at the COST WG2 Meeting ‘Putting halophytes to work – genetics, biochemistry and physiology’ Hannover, Germany, 28–31 August 2012.
Functional Plant Biology 40(9) 759-774 https://doi.org/10.1071/FP12285
Submitted: 27 September 2012 Accepted: 20 January 2013 Published: 4 March 2013
Abstract
The successful integration of activity in saline environments requires flexibility of responses at all levels, from genes to life cycles. Because plants are complex systems, there is no ‘best’ or ‘optimal’ solution and with respect to salt, glycophytes and halophytes are only the ends of a continuum of responses and possibilities. In this review, I briefly examine seven major aspects of plant function and their responses to salinity including transporters, secondary stresses, carbon acquisition and allocation, water and transpiration, growth and development, reproduction, and cytosolic function and ‘integrity’. I conclude that new approaches are needed to move towards understanding either organismal integration or ‘salt tolerance’, especially cessation of protocols dependent on sudden, often lethal, shock treatments and the embracing of systems level resources. Some of the tools needed to understand the integration of activity and even ‘salt stress’ are already in hand, such as those for whole-transcriptome analysis. Others, ranging from discovery studies of the nature of the cytosol to expanded tool kits for proteomic, metabolomic and epigenomic studies, still need to be further developed. After resurrecting the distinction between applied stress and the resultant strain and noting that with respect to salinity, the strain is manifest in changes at all -omic levels, I conclude that it should be possible to model and quantify stress responses.
Additional keywords: cation transport, chaperones, compatible osmotica, continuum, cytosolic integrity, glycophyte, halophyte, hydrophilins, photosynthesis, salt tolerance, signalling, sodium toxicity, water relations.
Introduction
All plants operate to accomplish a basic set of functions: resource acquisition, appropriate resource allocation to all individual cells and functions, and reproduction. Nearly all of the details of how they do this are open to negotiation, i.e. there are no universal solutions, because all organisms operate within constraints imposed by their own, unique and constantly changing environments. Moreover, the constraints operating through soil and aboveground environments vary with different spatial and temporal patterns. Successful integration of activity is dependent on flexibility of responses at levels from genes to metabolism to life stage progressions and the ability to adjust to, ameliorate or tolerate extreme or abrupt variations. As is clear from the diversity of plant species and forms persisting in virtually any ecosystem, there is no ‘best’ or ‘optimal’ solution.
Salinity is just one of many problems that plants have solved. Throughout this review, I will suggest that there are more fundamental similarities than differences between plants adapted to saline environments and those adapted to other conditions.
Salinity, like drought, has long attracted the attention of plant biologists. This interest has increased in the last 30 years, reflecting the fact that millions of hectares of land under cultivation with the most economically important crops have been adversely affected by salinity, usually as a result of inappropriate irrigation practices and other forms of land degradation (Munns 2005; Munns and Tester 2008; Zhang et al. 2010). A broad concern with how plants function in saline environments has been replaced with an almost singular focus on ‘salt tolerance’ of those plants which, on the whole, do not.
Since 2000, no fewer than 200 reviews have been published on ‘salt tolerance’. This paper will not be number 201. I will avoid simply covering the same ground again, instead espousing different, sometimes largely untested approaches to the broader problem of overall integration of activity. With respect to salinity, I am more concerned with success in predictable or slowly changing environments – whether they are daily and seasonal normal variations or the result of landscape level changes – than in the results of very short-term shock treatments. Given the prevalence of the latter in the current literature, this will necessitate a certain degree of speculation. However, given the complexity and intractability of the problem, I consider that justified.
Salt tolerance – finding the actual problem
Despite its immediate comprehensibility sensu ablato, i.e. in a completely informal structure, understanding salt tolerance has been an elusive goal. One major reason for this is that there is no quantitatively useful definition of ‘tolerance’, only situation-dependent proxies. For example, in breeding crops such as wheat or rice for ‘salt tolerance’, it is quite reasonable to measure growth or yield at different salinities or to define some scenario as a ‘control’ and compare individual performance to that. A similar approach is not useful for any long-lived or clonal perennial, however, and in either case, the results say nothing mechanistic about the differences in growth. Growth in annual crop breeding is simply a proxy for some perhaps Platonic ‘ideal’ of tolerance; it is not, however, a definition. Simply put, in attempting to study tolerance, there is nothing unequivocal to plot on a y-axis. However, because ‘tolerance’ is a concept both useful and important, I will explore in the final sections of this paper whether a generally applicable definition might be possible.
For the most part, considerations of plant–salt relationships have involved dividing the higher plants, or more specifically the angiosperms, into two groups, glycophytes and halophytes. Different approaches have been taken to define the strategies each group incorporates in dealing with salt, with by far the largest amount of attention being given to the glycophytes. It should be noted here that ‘strategies’ are only loosely structured summaries of what a plant or group of plants seems to do. They are not strategies in the more usual sense (e.g. business, sports, war); that is, they are not plans for the deployment of resources in order to achieve a goal. We have no nearly complete catalogue of the resources and for those which we can enumerate (e.g. the various putative ion transporters), we have very little information about their actual or potential deployment.
In glycophytes, the model for salinity tolerance centres around the declaration that sodium is toxic. ‘Toxicity’ is, however, another vague concept. Unlike cyanide, carbon monoxide, paraquat, or any number of other compounds whose mechanism of action can be defined, there is no target for sodium, only the knowledge that it is possible to kill a plant by putting salt in the soil. The most common explanation for sodium toxicity is that it has an inhibitory effect on activities of enzymes with little difference between halophytes and glycophytes (Flowers 1972; Greenway and Osmond 1972). Despite the frequent citation of these 40-year-old articles, it was unclear even in 1976 that this explanation was complete, even in vitro (Flowers et al. 1976; Jennings 1976). A similar conclusion was reached after careful re-examination in more recent reviews (Tester and Davenport 2003).
Nevertheless, because of its declared toxicity, in the glycophyte model, salt tolerance is dependent on excluding sodium from plants (Pardo and Quintero 2002; Munns 2005; Shabala and Cuin 2008; Plett and Møller 2010; Munns 2011). Even more critically, Na+ must be excluded from the shoot, and in all tissues, from the cytosol, although the nature of the cytosol itself is poorly understood (see below). In plants lacking sufficient control over Na+ fluxes to prevent shoot accumulation, salt build-up in leaves leads to their mortality and eventually to that of the entire plant (Munns and Tester 2008; Munns 2011). As I noted 25 years ago, ‘Although the processes of physiological folklore have elevated [sodium toxicity] to a belief in the almost paranoiac avoidance of cytoplasmic Na+, it is not at all clear what levels of Na+ are actually biochemically unacceptable’ (Cheeseman 1988). This remains the case.
Clearly, little of the basic model applies to halophytes despite efforts to stretch it that far (e.g. Yadav et al. 2011). In halophytes, tissue Na+ concentrations can easily exceed 500 mM and there is considerable evidence against its extreme exclusion from the cytosol (Flowers and Colmer 2008; Kronzucker and Britto 2011). External Na+ levels that would seriously reduce growth rates or kill glycophytes may stimulate or even be required for maximal growth of halophytes. Indeed, Na+ may stimulate both K+ uptake and growth (Flowers et al. 1977; Flowers and Colmer 2008). Moreover, despite the sometimes high leaf tissue concentrations and continued transpiration even under seawater conditions, salt does not, in fact, accumulate to toxic levels in older leaves even with continuous and sometimes direct exposure to high external concentrations (e.g. Cram et al. 2002). Limiting energy supplies for transport by reducing photosynthesis also does not lead to sodium accumulation or ‘toxicity’ (Cheeseman and Wickens 1986).
The beneficial effects of sodium for plant growth are also not limited to halophytes (Flowers and Läuchli 1983; Subbarao et al. 2003). It has been known for a long time that many other plants benefit from the availability of salt to varying degrees (e.g. von Marilaun 1896; p. 74). For more than 150 years, the Rothamsted Park Grasslands experiment has applied different fertiliser treatments to experimental plots; nitrate is applied as a sodium salt (Silvertown et al. 2006).
From this, it can be concluded that the model on which ‘salt tolerance’ research is based is vague at best. A new approach is needed that focuses on careful and quantitative dissection of both global and individual processes and the more complete problem of integration from the cellular to the organismal level over a range of soil conditions. That is the subject of the rest of this paper.
Glycophytes, halophytes and the sodium content continuum
The first step in redefining the problem is to recognise that ‘glycophyte’ and ‘halophyte’ are simply convenient labels given to plants at the extremes of a continuum. Conceivably, this continuum should be tabulated as the maximum salinity at which a species could complete its life cycle, but that is too dependent on other conditions to make its determination feasible and may be impossible in clonal species and long-lived perennials. Alternately, it can at least be demonstrated based on leaf salt concentrations of ‘healthy’ plants, i.e. ones not clearly in the process of dying, under controlled or natural conditions. In that case, this continuum has been clear since the first, laborious analytical methods for distinction between Na+ and K+ were established (Bertrand and Perietzeanu 1927; Lundegårdh 1929). Using Lundegårdh’s new flame emission spectrometer, Collander (1941) reported Na+ contents for 21 species under controlled conditions, with values ranging from 0.5% of the total monovalent cations in Fagopyrum, to 30% in Atriplex hortense; there were no gaps in the continuum. More recently, the continuum is also at least implied by the relative growth responses to NaCl and their classification into four generalised responses (summarised by fig. 1 by Greenway and Munns (1980)).
There is also, of course, a continuum in the relative ‘salt tolerance’ of crop plants, as shown by the classification of 65 crops by Yadav et al. (2011). Even wheat and rice show considerable variation between genotypes (Ali et al. 2006; Munns and Tester 2008) and in tetraploid wheat (Triticum turgidum), this variability is, significantly, not correlated with the extent of Na+ exclusion (but see Munns and James 2003; Genc et al. 2007). A poor correlation between Na+ exclusion and tolerance was also reported for rice (Yeo et al. 1990). Arabidopsis thaliana, often considered a model glycophyte, shows considerable variation within the species (Trontin et al. 2011). Moreover, it fails to show the expected glycophyte-associated correlation between Na+ accumulation and salt sensitivity; mutations which increase sensitivity do not necessarily show increased shoot Na+ accumulation (Essah et al. 2003; Tester and Davenport 2003).
Viewed this way, the integration of activity in saline environments should be considered as a very interesting set of problems to be analysed in a variety of species, not as a limited set of contrasts between ‘halophytes’ and ‘glycophytes’.
The problem of organismal integration in saline environments can be solved in many ways
Thirty-five years ago, Flowers et al. (1977) reviewed the physiology of halophytes. Their article still provides an excellent foundation for understanding organismal options and particularly for addressing apparent differences between species. Moreover, their discussion applies to plants across the continuum. Briefly, they reported that studies of halophyte growth at high concentrations of salts ‘offer a rather bewildering array of data’. As halophytes occur in at least 37 orders of plants, (see also Flowers et al. 2010), this should not be unexpected. Optimal growth salinities range from 20 to 500 mM with large differences even within single genera (e.g. Atriplex). This can vary considerably with life history stage. The addition of low levels of NaCl (e.g. 1–10 mM) often results in dramatic increases in growth. In this regard, as well as in internal levels of salts which are ‘toxic’, species differ considerably in their responses to Na+, K+, Mg2+ and Ca2+. The nature of the anion may or may not make a difference. Potassium uptake may or may not be stimulated by Na+; the stimulation may be greatest at K+ levels typical of non-saline environments rather than those of their native habitats. Indeed, K+ uptake may decline more severely when Na+ is added to solutions with high (50 mM) K+ than those with low K+.
Solutions to the problems of adaptation to salinity must be embedded in the genome. The ‘bewildering array’ of characteristics summarised by Flowers et al. (1977) undoubtedly reflect the multitude of ways in which genomes evolve and the potential for rapid adaptation to environmental conditions (Oh et al. 2012). These need not precede speciation. In some cases, it has been shown that evolution – especially through tandem gene duplication – proceeds more rapidly in plants experiencing stress and that the duplications can be retained for long times (Dassanayake et al. 2011; Oh et al. 2012). With respect to salinity, following the sequencing of the genomes of two halophytic Arabidopsis relatives (Thellungiella parvula and T. salsuginea), it became evident that genes associated with abiotic stress responses and ion transport had been tandem duplicated in the halophytes in significantly higher numbers than in Arabidopsis (Dassanayake et al. 2011; Oh et al. 2012; Wu et al. 2012). These duplications, although showing a high degree of sequence similarity to their Arabidopsis equivalents, have essentially no similarity in their promoter regions and substantially different expression patterns even in the absence of salt (Oh et al. 2010). How their epigenomes or populations of small interfering RNAs differ has yet to be examined, but these are undoubtedly involved in their different responses to stressful environments.
Crop species also show high variabilities in gene expression in response to salinity. A recent study by Walia et al. (2009), for example, compared gene expression patterns in rice, barley and wheat roots. Between rice and barley or rice and wheat, they found no major conserved whole transcriptome expression patterns. In the more closely related barley and wheat, only about one-third of the salinity-related expression responses were overlapping. Intraspecific variation in gene expression is also manifested in locally adapted populations of model plants (Baxter et al. 2010; Friesen et al. 2010).
Major aspects of plant function and their relationships to salinity
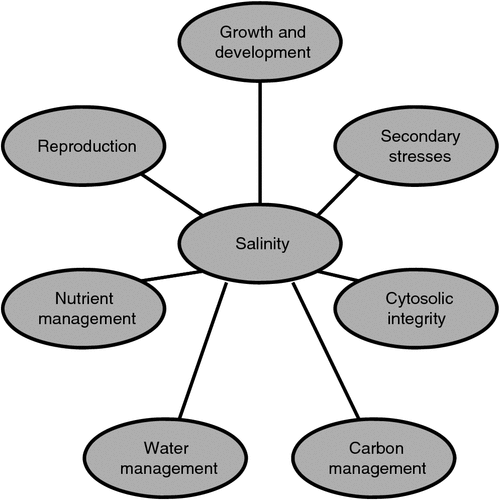
Figure 1 illustrates schematically seven major aspects of plant function which must be integrated for a plant to succeed in a saline environment. Clearly, the central bubble (salinity) can be replaced with any other environmental factor and the diagram is still applicable. The least well understood of these interactions is that of salinity and the control of reproduction. This problem is now beginning to be addressed at the molecular level with studies such as that by Kim et al. (2013). These authors reported that in A. thaliana the flowering time regulator, GIGANTEA (GI), physically interacts with the SNF1-related protein kinase, SOS2 under non-saline conditions. This interaction prevents or reduces SOS2-based SOS1 activation. Under saline conditions, GI degrades, releasing SOS2. The sum effect is a delay of flowering and at least a partial maintenance of growth (i.e. increased ‘tolerance’). Hopefully, this study will lead to better explanations of higher-order molecular interactions associated with growth characteristics, perhaps including how reduced growth and reproduction are adaptive under saline conditions.
|
Transporters
Undoubtedly, transporters have received the greatest emphasis of any of the interactions in Fig. 1. Since 2000, there have been 69 reviews of salt tolerance which have focussed on the monovalent ion transport problem, so a detailed recapitulation is not needed. Overall, at least six different possible mechanisms have been identified which putatively move Na+ and/or K+ across the plasma membrane and the tonoplast. These are the non-selective cation channels (NSCCs), especially the voltage insensitive or VI-NSCCs, one or more ‘low affinity’ cation transporters (e.g. LCT1), two families of predominantly K+ transporting enzymes (KUT/HAK/KT and AKT), two families of ‘high affinity’ transporters (HKT1 and HKT2), at least the former of which predominately carries Na+ (see Horie et al. 2009) and at least two neutral antiporters (SOS1, for Na+/H exchange at the plasma membrane and NHX-family proteins catalysing exchange at the tonoplast). The status for each of these groups was reviewed recently by Kronzucker and Britto (2011) who raised serious questions about their proposed individual functions. Moreover, none has yet been integrated into a model elucidating their deployment and functioning in an organismal context.
The importance of maintaining a low cytosolic Na+ concentration has been agreed upon to the point of being dogma, at least partly on the evidence that animal cells depend for their continued existence on maintenance of low Na+ concentrations. But animal cells do not have central vacuoles amounting to 95% of their volume and they do have sodium extruding ATPases. The apparent biophysical aspects of the balance cannot be lightly discounted (e.g. Carden et al. 2003); the inward electrochemical gradient on Na+ at the plasmalemma, the tendency of Na+ to move down the gradient (Cheeseman et al. 1985; Xue et al. 2011) and the tendency of Na+ to cycle rapidly, make it difficult to reconcile measurable fluxes with the energy available from respiration (Malagoli et al. 2008; Britto and Kronzucker 2009; Kronzucker and Britto 2011). In addition, even at moderate salinity in both glycophytes and halophytes, there can also be a substantial electrochemical gradient across the tonoplast driving Na+ back to the cytosol (Carden et al. 2003). This exacerbates the energy problem; it is not simply a problem at the plasmamembrane. Clearly, there is still something critical we are missing about the status of Na+ in cells (Lazof and Cheeseman 1986).
However, the problem is not simply limited to individual cells in their generic sense. For example, Tester and Davenport (2003), although noting the importance of the rapid influx and turnover of Na+, also noted that cells in different parts of roots (or shoots) have to behave very differently with respect to transport in order to bring about the coordinated distribution of Na+ and to prevent excess accumulations. Moller et al. (2009) subsequently demonstrated this elegantly with their study of HKT1;1 in A. thaliana. If this differential behaviour is absent, any ion that is delivered to the xylem in the roots will appear in the shoot if it is not somehow removed along the way (reviewed by Karley et al. 2000). Once there, it has to be either accommodated or recirculated (Jacoby 1979; Berthomieu et al. 2003; Kong et al. 2012) or in plants with salt glands, partly excreted (Flowers et al. 2010).
The problem is easy to envisage: consider a plant with large, older leaves and small, young ones. As noted by McNeil et al. (1979) in their studies of amino acid distribution in lupins, the larger leaves get the bulk of the transpirational water, but they generally do not need the bulk of the nutrients. Instead, there is a transfer from xylem to phloem or xylem to xylem at the departing leaf traces, redirecting amino acid flow. With the possible exception of species which accumulate Na+ in older leaves to an extent which causes necrosis (e.g. rice Yeo et al. 1985), a similar transfer system must exist for that ion. As noted earlier, even in halophytes under seawater conditions, transpiration continues, but salt accumulates in older leaves only slowly and not to toxic levels. Indeed, the concentrations in young leaves are nearly as high as those in older leaves (Cram et al. 2002).
Thus, although the long distance transport of ions cannot be separated fully from consideration of water flows, the fates of water, ions and other nutrients in the transpiration stream are largely independent. The study of mechanisms and controls of long distance transport must still be considered to be in its infancy, but it is difficult to see how any list of genes can be translated into understanding of organismal strategies without expanding this considerably. Park et al. (2008) outlined a series of criteria that could be used to distinguish transporters involved in long distance transport. At the organismal level, these included, in particular, demonstration that expression was concentrated in the vascular tissue system and that mutants and wild-type plants had different shoot ion concentrations and altered phloem or xylem concentrations. Based on this, only a few transporters could be implicated as involved for K+ and Na+ (SKOR, SOS1 and HKT1 and HKT2) (see also De Boer and Volkov 2003; Maathuis 2007). In no case, however, was there enough evidence to suggest how strategic, cell or tissue deployment of the proteins could be integrated with observations of overall ion distributions.
From the 1950s to the ’80s, there were countless studies of transport physiology at the tissue and organismal level. These were largely dismissed as phenomenological once the molecular (‘mechanistic’) era began in the 1980s. However, models for cell or tissue deployment of individual transporter proteins that can quantitatively, or even qualitatively, explain any single ‘phenomenological’ study have been very slow in coming. To the best of my knowledge, the only example so far comes from the recent work on HKT1, largely from Tester’s group. They have elucidated many of the molecular, biochemical and physiological ramifications of its expression and tissue localisation (Davenport et al. 2007; Moller et al. 2009; Munns et al. 2012) and recently presented a two-stage, 3D model based on the crystalline structure of various HKT1 isoforms that provides an explanation for Na+ exclusion in rice (Cotsaftis et al. 2012). To make significant progress here, many more such studies combining physiological, genetic and biophysical approaches will be needed.
Secondary stresses
The primary concern with secondary stresses associated with salinity (or drought) has been with oxidative stress following salt shock. Analyses have generally been limited to a small number of enzymes (superoxide dismutase, ascorbate peroxidase, glutathione reductase, catalase, phenolic peroxidases and NADPH oxidase), to estimates of membrane oxidation and membrane leakage and, with varying degrees of reliability, to the measurement of H2O2 and other oxidants (Cheeseman 2007). This list has not changed in two decades. Overall, there has been very little attention paid to oxidative enzyme activities or oxidant levels as normally occurring and to their positive and essential roles in growth and development (see Cheeseman et al. 1997; Cheeseman 2006, 2009).
At the molecular level, there has been considerable progress in understanding the responses of gene expression associated with oxidative signalling pathways following shock treatments (e.g. Iglesias et al. 2010; Miller et al. 2010; Golldack et al. 2011). This work has yet to be incorporated in models of the overall integration of activity under even the most benign conditions, much less in the more challenging case of saline environments (Cheeseman 2007).
Carbon acquisition and allocation
It is impossible to consider organismal integration in any environment without taking photosynthesis and carbon metabolism into account. Carbon is the major shoot-derived resource and the source of all energy expendable to support metabolism. Surprisingly, however, many studies, in particular those using Arabidopsis, use plants grown at too low light levels to sustain growth, instead substituting root-medium sucrose as the carbon source. The majority of other studies, if they deal with photosynthesis at all, usually report CO2 exchange, stomatal conductance and the other basic parameters reported by any infrared gas analyser system, presenting them as dependent variables varying with time or salinity. Overall, it is difficult to say that such studies provide anything more than an intriguing suggestion that photosynthesis is related to (in this case) salinity.
In contrast, a small number of sentinel papers have explored the problem more deeply. For example, the relative limitations on photosynthesis due to diffusive and metabolic factors were carefully analysed by Flexas et al. (2004), who concluded that until stomatal conductance fell to below 0.1 mol m–2 s–1, the limitations were almost entirely stomatal, i.e. metabolic changes did not occur until more serious stresses were developed. With respect to plants adapted to saline environments, this threshold conductance may actually be towards the higher, not lower, end of the in situ range (e.g. Cheeseman et al. 1991; Cheeseman and Lovelock 2004) and in those conditions, such manipulations as increasing external CO2 to very high levels to overcome stomatal limitations, have generally not led to dramatic increases in net assimilation rates. Something other than stomatal conductance is limiting photosynthesis rates.
Photosynthetic performance is itself a complex systems problem (Cheeseman and Lexa 1996). Performance is modified in many ways in saline environments including effects on chloroplast functioning per se, i.e. on the light dependent and independent reactions (Chaves et al. 2009, 2011). Stepien and Johnson (2009) detailed many of these in a comparative study of Thellungiella salsuginea (halophila) growing successfully at salinities up to 500 mM NaCl and A. thaliana failing to thrive at 100 and 150 mM NaCl. Over a period of 2 weeks, both net assimilation and stomatal conductance declined in A. thaliana, but showed little if any change in T. salsuginea. By 10 days of salinisation, the dependence of net photosynthesis on intercellular CO2 (A.Ci) for A. thaliana leaves showed loss of both carboxylation efficiency and Vcmax, whereas fluorescence analysis showed loss of maximum photochemical efficiency of PSII (Fv/Fm) and Fv′/Fm′, the maximum efficiency of PSII in the light. In contrast, even at 500 mM NaCl, T. salsuginea showed no compromises in performance of either electron transport or Calvin cycle components. The amount of energy dissipated as heat in A. thaliana chloroplasts (non-photochemical quenching) also increased, particularly the slowly recovering component associated with photoinhibitory damage. With regards to PSI activity, with time P700 became more oxidised in A. thaliana and overall electron conductance through the whole chain declined.
With the Calvin cycle compromised or downregulated, the importance of alternate methods of consuming electrons increases, especially those associated with photorespiration, photoinhibition (xanthophyll cycle) and delivery of electrons to molecular oxygen (in particular, the Mehler reaction). In some cases, such as wheat and barley under drought stress, photorespiration plays the major role in dissipation of excess energy (Noctor et al. 2002), but this is not uniformly the case. In both A. thaliana and T. salsuginea, photorespiratory O2 fixation was unaffected by salinity (Stepien and Johnson 2009). In A. thaliana, increased cyclic electron flow around PSI accompanied salinisation, whereas in T. salsuginea, electrons were dissipated instead by a plastid terminal oxidase (PTOX) (Stepien and Johnson 2009).
The problem of excess electron transport capacity can also be solved by detoxifying the products of the Mehler reaction using the antioxidant enzymes superoxide dismutase, chloroplast ascorbate peroxidase and glutathione reductase (Asada 1994; Allen 1995; Cheeseman et al. 1997). The extent to which this happens is difficult to assess because the activities for the essential enzymes are almost always expressed on a relative basis. Cheeseman et al. (1997), however, using an oxygen electrode method for SOD determination, showed that the mangroves Rhizophora stylosa and R. mangle had SOD activities more than sufficient to consume all the electrons produced in the light dependent reactions and thus would likely be able to convert any Mehler reaction derived •O2– to H2O2 efficiently enough to prevent photobleaching.
The potential H2O2 production was, however, up to 40 times higher than the activity of the next enzyme in the detoxification process, ascorbate peroxidase. One reasonable explanation for subsequent H2O2 removal was by a highly active, heat stable, phenolic peroxidase (Pearse et al. 2005). Nevertheless, in general, the potential for over reduction of the cytosol remains, associated with high rates of NADPH production and low rates of CO2 or O2 fixation (Haslam 1986). To maintain a balance between photosynthesis and productive uses of C (i.e. growth) in rhizophoracean mangroves when nutrients are seriously growth-limiting, and to prevent over-reduction of cytosolic metabolic pools, more than 50% of recently fixed C is diverted to tannins, flavonoids and other metabolically expensive (ATP consuming) products of the phenylpropanoid pathway (Kandil et al. 2004). Production of anthocyanins (in other species) may also be a response to such imbalances. Another mechanism for dissipation of excess NADPH reducing power has been reported in celery, i.e. the upregulation of mannitol synthesis driven by mannose-6-phosphate reductase (Everard et al. 1994).
Water and transpiration
Since the development of the ‘pressure bomb’ for measuring xylem water potentials and its application to the study of trans-root water movements in mangroves (Scholander 1968), it has been known that water and ion movements across roots are nearly completely uncoupled, that is, that there is an ‘ultrafiltration’ system separate from the suberised Caspian strip which restricts water-driven ion movements (see also Läuchli et al. 2008). This is true not only for Na+, but for any ion in any soil solution, even if it is obvious only for those which are present at levels in excess of what can be used at exactly the rates of bulk water delivery.
It is now apparent that no matter the plant or the environment, control of water movements resides largely in the activity and specificity of aquaporins, especially those at the root–solution interfaces (Maurel et al. 2008, 2009). An exception may be the case of by-pass flow in rice (Yeo et al. 1987; Faiyue et al. 2010).
Recent research has established not only that aquaporin gene families are large (~35 in A. thaliana and approximately the same number in the halophytic Thellungiellas), but also that the individual proteins and their placement in membranes is highly dynamic. The bulk of this work has been done using A. thaliana and short-term shock treatments. As reviewed recently by Aroca et al. (2012), typically, but not universally, PIP and TIP transcript levels decrease upon shock treatment with salt or drought and recover over a period of days as plants acclimate to their new conditions. The decline in the associated proteins does not, however, simply reflect degradation, but re-localisation to intracellular structures (Boursiac et al. 2005). Studies of PIP1;2 and PIP2;1, in particular, have found that dispersion is heterogeneous and that salinity stress increases their internalisation and reduces their presence in the plasma membrane (Li et al. 2011; Luu et al. 2012). The subcellular trafficking is mediated by both clathrin and membrane raft pathways.
Overall, at least five PIPs in A. thaliana form a co-expression network regulated by a duplicated pair of DREB transcription factors, RAP2.4B and RAP2.4 (Rae et al. 2011). The latter was uniquely induced by salinity treatment. Salinity-induced aquaporins have also been reported in other species, such as Leymus chinensis, a grass species indigenous to sodic alkali habitats (Ma and Liu 2012) and hybrid poplar (Bae et al. 2011). Salt induction of aquaporins does not seem to be a ubiquitous phenomenon, however; in citrus, NaCl did not affect PIP1 or PIP2 transcript levels even though root hydraulic conductivity and transpiration were both reduced (Rodríguez-Gamir et al. 2012).
I suggest that although the water relations of plants in any environment, saline or otherwise, have been studied for more than 50 years, a different and more mechanical understanding is now closer to hand. It is important, however, not to approach this simply by transferring single aquaporin genes from one species to another in hopes of finding the one that confers a miraculous level of drought or salt tolerance on the target. Useful results taking that approach are likely with exceedingly low probability and little of mechanistic interest will likely emerge as a by-product. An alternative approach based on modulating expression of multiple aquaporins using RNAi may be possible; Sangster et al. (2004, 2008a, 2008b), for example, used this approach successfully to determine the function of the HSP90s in A. thaliana.
Growth and development
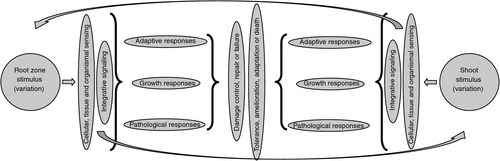
Figure 2 illustrates a basic schematic model for response of a plant to any environmental stimulus. Until recently, all that could be done was to apply a stimulus and observe far-downstream growth, physiological or pathological responses. The more detailed the observations and measurements, the greater the detail that can be filled in to the bits in the middle of the figure. The problem changes, however, with each phenological stage, i.e. with each life stage and environmental sieve that is passed.
|
With respect to nutrients in particular, plant responses are both nutrient and species specific (López-Bucio et al. 2003; Lambers et al. 2008). All measured responses also depend heavily on conditions prevailing or varying throughout the growth period. Such peripheral influences include such factors as salinity, water, temperature and light, as well as stochastic root or shoot proliferation events (‘developmental instability’ Forde 2009). In the longer term, there is also considerable cross talk between metabolic processes at all levels of spatial and temporal organisation (Schachtman and Shin 2007).
Historically, the most critical and difficult aspects of the whole response network in Fig. 2 have been those associated with perception and signalling (Hirayama and Shinozaki 2010). It has also been problematic to distinguish responses which reflect primary sensing and signalling from those which represent general or non-specific stress responses (Hammond et al. 2003). Root activity involves sensing the environment and adjusting transport systems and root proliferation as necessary (Walch-Liu et al. 2005). Balanced organismal-level responses depend on integration via local and systemic signalling (Schachtman and Shin 2007). Although individual roots respond to local conditions, integrated responses are systemic, i.e. involving the shoot and roots that may themselves not be under a nutrient limitation.
Knowledge of individual sensing and response networks is now emerging based on large scale transcriptomic studies using response time-courses and model plants. These involve multipoint measurements of gene expression using microarrays or RNAseq, detailed quantification of hormone and metabolite levels and fluxes and detailed measurements of particular aspects of growth. For these, the split root system approach has proved useful in differentiating between local and systemic responses (Attia et al. 2008; Bazihizina et al. 2012).
The response to nitrate supply is the best example, to date, of the application of transcriptomic studies to split root systems, but advances have also been made with respect to phosphate, potassium, iron and sulfur (Amtmann et al. 2006; Schachtman and Shin 2007). Using A. thaliana, Ruffel et al. (2011) have documented the transcriptomic changes underlying local responses to nitrate availability, albeit under Petri dish, heterotrophic growth conditions. These responses are regulated by the nitrate and auxin transporter/receptor (hence, ‘transceptor’) NRT1.1 (At1g12110) in a network which also includes ANR1, NLP7 and SPL9 transcription factors and kinases CIPK8 and CIPK23. Although long distance signalling is less completely understood, they found that amino acids (especially Glu/Gln) provide feedback that prevents lateral root outgrowth in a network that involves auxin response factors (ARF8) and miR167. Hormones also play a role in signalling of nitrate status, especially cytokinins (CK) as evidenced by the fact that (i) NO3– supply induces IPT3, which is the first enzyme in CK biosynthesis and (ii) CK regulates expression of N uptake and assimilation genes as well as root architecture. Ruffel et al. (2011) established that CK is indeed a root-to-shoot signal molecule and a component of the signalling/relay mechanism carrying information about the NO3– status of the plant, enabling root growth and improving foraging when NO3– is limiting. Auxin produced in the apical meristem of the shoot plays an antagonistic role in the root, thus further refining the overall response.
The use of microarrays and, more recently, RNAseq techniques in such studies has great potential for both individual gene- and systems-level understanding, although the massive amount of data which results has the potential to make specific interpretations difficult (e.g. Seki et al. 2002; Chao et al. 2005; Walia et al. 2006). The greatest strength of the study by Ruffel et al. (2011) was the use to which they were able to put transcriptomic analysis using microarrays. In all, expression of 123 genes was altered in the interaction between N availability and the split root condition. The clustering of the expression of these genes in the different tissues was similar to the lateral root growth patterns of the treatments. Krouk et al. (2010) were able to extend the nitrate study to the next level by using multiple time points and modelling short-term responses.
It is clear, however, that with increasing numbers of repeats, treatments and tissue types come better models. Ma and Bohnert (2007) for example, analysed all of the available A. thaliana microarray data to 2007 to identify transcripts responsive to salt stress (i.e. sudden shock), identifying 1500 genes which were strongly regulated by the treatment. Although the majority responded to other abiotic stresses as well, 171 transcript responses were specific for salt treatment, mostly in the roots. Of these, nearly 20% were transcription factors, but there were also 35%, which were ‘unknowns’. Similarly, Seki et al. (2002) analysed the responses of 7000 Arabidopsis genes under abiotic stress (shock) conditions, associating 89 with response to NaCl, but only 34 of these were restricted to salt.
It is now only a matter of time before the networks for model systems are extended even further to include more detail about hormone and metabolome interactions (Van Norman et al. 2004) and about specific things which need to be accomplished during the growth process, such as vascular development (Dettmer et al. 2009) and the critical roles played by shoot and root apical meristems (Galinha et al. 2009). Given this progress, the stage is now certainly set to use similar techniques to advance our understanding of responses to salinity and their effects on other processes (Bazihizina et al. 2009, 2012).
Sodium toxicity, cellular water and cytosolic conditions
Organismal integration in saline environments depends on a sophisticated system for controlling the distribution of sodium, but in each tissue, the ultimate point of integration is at the cellular/cytosolic level; if it fails here, then death results. This is also the level at which water relations, osmotic balance and specific ion effects intersect. Arguably the most critical requirement is maintenance of an appropriate level of protein hydration in the cytosol. In this section, therefore, I will look at some of the overlapping functions involved in maintaining an acceptable cytosolic milieu.
The cytosol is very poorly characterised in plants. However, based on studies primarily of bacteria, it is first and foremost, neither a dilute aqueous solution nor a sack of enzymes and metabolites interacting by random diffusion. It is, instead, highly crowded (Verkman 2002; Spitzer and Poolman 2005; van den Bogaart et al. 2007; Spitzer and Poolman 2009). When biomacromolecules occupy 20–30% of the cytoplasmic volume (the normal situation) it becomes highly structured with spatially and temporally self-constructing molecular devices. The crowded regions act, effectively, as three dimensional membranes permeable to cations and conditionally permeable to anions. Both screened and unscreened electrostatic forces play important roles in the regulation of metabolism and transport. This crowding was probably essential to the evolution of life and is essential for its continuation (Spitzer and Poolman 2009).
Despite the fact that they are difficult to study and not identifiable at the gene or transcript levels, the existence of large molecular devices (multiprotein complexes) in the cytosol is very well established (Wiggins 1990; Book et al. 2010; Olinares et al. 2010). Indeed, it is possible that in an intact system, there are few ‘soluble’ enzymes; they only appear when cells are disrupted and their contents diluted. Such complexes include a wide variety of cellular machines such as the kinetochore (Welburn and Cheeseman 2008) and the complexes responsible for DNA replication and RNA translation. In addition, there are membrane complexes such as clathrin (Batchelder and Yarar 2010) and multifunctional enzymes in primary metabolism (e.g. pyruvate dehydrogenase, Behal et al. 1993; Perham 2000), the Calvin cycle (Wedel et al. 1997), the phenylpropanoid pathway (Winkel 2004), glycolysis (Graham et al. 2007), polyamine biosynthesis (Alcázar et al. 2011) and starch synthesis (Hennen-Bierwagen et al. 2009; Liu et al. 2009). Although some of the complexes ‘make sense’ based on well-known biochemistry, others, such as the association of aldolase with the vacuolar ATPase, are less obvious (Lu et al. 2004). In this case, however, aldolase interacts with the a, B and E subunits of the V-ATPase (Lu et al. 2004), coupling the ATP-generating glycolytic pathway with the ATP-hydrolysing proton pump.
Enzyme kinetics and binding equilibria typically depend on ionic strength in complex ways. Some of the dependencies are for specific ions, especially cations (e.g. K+, Na+ or Mg2+) and some are general (Suelter 1970; Clarkson and Hanson 1980; Nayal and DiCera 1996; Page and Di Cera 2006). Some of these dependencies that are evident in vitro disappear in vivo, possibly reflecting the consequences of condensation of enzymes into large complexes, i.e. to macromolecular crowding. This difference is quite probably responsible for the concept that enzymes are, in general, much more sensitive to Na+ than K+ (in both halophytes and glycophytes, Greenway and Osmond 1972) and that Na+ is toxic. This, in turn, has led to the dogma that Na+ must be excluded from the cytosol despite evidence to the contrary (e.g. Table 1 in Flowers and Colmer 2008).
The ionic conditions of the cytosol affect metabolism in several ways. For example, cyclic metabolic and replicative reactions can only occur in the cytosol within certain ranges of stabilising non-covalent forces as determined by ionic strengths (Spitzer and Poolman 2009); interestingly, when these are studied in vitro, ionic strength is typically adjusted using NaCl. The stabilising forces are determined in part by the distance from a particle or surface over which mobile charge carriers can screen out the external electric field, i.e. the Debye’s length of charges at macromolecular and membrane surfaces. This distance increases rapidly at ionic strengths below ~0.1 M but becomes small, acceptably less than 1 nm, for a wide range of concentrations above that. Spitzer and Poolman (2009) noted that these concentrations/Debye lengths are commensurate with the surface-to-surface distances of macromolecules occupying 20–25% of cell volume and conclude that crowding and short Debye lengths were necessary for the emergence of dynamic vectorial pathways and structures.
The fixed charge and ionic conditions of the cytosol inevitably determine the local water relations relevant to the cytoskeleton and proteins (Wiggins 1990). Critical differences are reflected in solvent properties, densities and mobilities. The equilibrium distribution of water within cells depends on uniform chemical potentials (water potentials) throughout a cell. These are functions of water activity, molar volume, pressure and temperature. Given that the latter two are constant throughout a cell, if the activities of water in different regions differ, e.g. because of differences in numbers of solute molecules, then the molar volumes have to change reciprocally. Thus, water equilibrates by increasing in density where solute concentrations are high and decreasing in density where concentrations are low. At the molecular level, these changes reflect the organisation of water, i.e. the numbers and linearity of the H-bonds between water molecules. Low density water is able to form more stable H-bonds and thus, its mobility is also reduced.
These differences, in turn, alter the partitioning of ions within cells (Wiggins 1990). Within a cell characterised by local regions of high density charge (protein and membrane surfaces), small, highly hydrated ions (Mg2+, Ca2+, H+ and Na+) tend to accumulate in the regions of greater density, while larger, singly charged ions (K+) are preferentially found in the less dense regions. Thus, Na+ and K+, which behave similarly in dilute aqueous solutions, partition in opposite manners in cells. For charge screening, however, the cations involved tend to be monovalent and depending on availability, different species may be involved to different extents. Clearly, if Na+ is not available (as in some low salt growth conditions), the screening role will be played by K+ even though the size and charge density of Na+ make it better suited (Wiggins 1990).
This alone may explain why plants growing in the absence of Na+ at significant levels have to have high cytosolic K+. With even low levels of Na+ availability, some of this K+ can be released for other purposes, e.g. for K+-specific protein activation (Clarkson and Hanson 1980; Flowers and Läuchli 1983). In the usual way of calculating ‘use efficiency’ as the inverse of tissue concentrations, potassium use efficiency increases along with decreased uptake (Messedi et al. 2004). This once broadly recognised ‘potassium-sparing’ effect of Na+ has, seemingly, been reinterpreted (or misinterpreted) as a deleterious effect of Na+ on transporters associated with determining ‘selectivity’. ‘Nutritional’ or functional (Subbarao et al. 2003) Na+ uptake under low K+ or K+-starvation conditions has even been identified in rice, one of the more salt-intolerant model species, and associated with OsHKT2;1 (Horie et al. 2007).
Returning to the status of water in cells, if small ions are the only osmolytes in the cells, the regions between the polyelectrolytes will, to varying degrees, be depleted of those ions (and osmotica). To maintain equality of water potentials between the regions, the cytoplasmic water (i.e. that not associated with the polyion surfaces), has to decrease in density, becoming more organised and less mobile. The compounds usually referred to as ‘compatible osmotica’, such as glycine betaine, proline, or sorbitol, are excluded from the regions near the polyions by their size and charge (or lack thereof). They will accumulate, instead, in regions with higher density water and in the process, decrease it (Wiggins 1990). Thus, compatible osmotica may well not play a chaperone-like role or protect the polyions from the evil machinations of Na+. They may, nevertheless, influence the overall water behaviour within cells.
There are, however, unquestionably conditions in which proteins must also be protected against denaturation and in which re-folding after stress is critical. This is accomplished by protein-based modulators of protein surface chemistry in the cytosol, i.e. the chaperones (heat shock proteins, Csermely et al. 2007) and the intrinsically disordered proteins (a.k.a. hydrophilins, LEAs, dehydrins or CORs, Battaglia et al. 2008). Unfortunately, the exact functions played by both groups for plants under stress and their mechanisms of action, remain to be fully characterised.
The roles and actions of chaperones include four critical tasks. These are: (1) local dissipation of metabolic noise, whether it originates with extracellular conditions or intracellularly; (2) efficient and reliable signal transmission; (3) distinction between signals and noise and the continuous remodelling of the networks as conditions change; and (4) protection against continuous random damage, such as that which may arise from free radical attacks (Csermely et al. 2007). Chaperones also buffer genetic variation, meaning that there are several naturally occurring phenotypes that remain unseen, or ‘cryptic’ until their activity is impaired, either by mutation or by environmental stress (Sangster et al. 2004). This chaperone-dependent natural variation appears to be common; in fact, Sangster et al. (2004) could reasonably suggest, for HSP90s, that there is at least one such polymorphism for nearly every quantitative trait (Sangster et al. 2008a). With the growth in popularity of ‘systems biology’, it should be easily recognised that none of these can be understood without reference to all the others, i.e. the integrated operation of cells (and organisms) are ‘emergent properties’ of the system.
Unlike the chaperones, which require ATP for their activity, the hydrophilins or intrinsically disordered proteins effect stress tolerance without direct energy input. Hydrophilins include the late embryogenesis abundant (LEA) proteins, the first group having been discovered in studies of seed desiccation. Other names for the group (or subgroups) are dehydrins and cold regulated genes (COR). The LEA hydrophilins are further divided into seven diverse groups, but share the characteristic of being largely devoid of secondary structure (random coil in solution) except when drying or in the presence of sugars or various other small solutes (Battaglia et al. 2008). In that case, they may form α-helices and intermolecular extended β-sheet structures. These structures, however, fully revert to the random coils upon rehydration.
Because the hydrophilins are highly charged, hydrophilic and have a high degree of hydration, they can interact with membranes, membrane proteins and protein complexes, protecting them against dehydration. Despite their size, their geometry and distribution of fixed positive charges allows them to interact with the fixed negative charges on other proteins. Hydrophilins can function to protect enzyme activities (and thus, probably also the conformation of other enzymes and enzyme complexes) during partial or nearly total desiccation, not only resulting from water withdrawal but by imposition of ionic and temperature stresses. Also unlike the chaperones, they do not facilitate re-folding of denatured proteins.
Moving forward
Over the past decade, advances in genomics, transcriptomics, epigenetics, proteomics and metabolomics have ushered in a new era, now broadly known as ‘systems biology’, bringing with it optimism for technical solutions to problems associated with salt and other biotic and abiotic stresses (e.g. Zhu 2001; Apse and Blumwald 2002). Although the optimism continues, at least guardedly, it has now been tempered by the realisation that ‘in the field, plants are subjected to various stresses simultaneously and the constraints (may) extend throughout their lifetimes’ (Hirayama and Shinozaki 2010). In their assessment of ‘the way forward’ in breeding crops for salt tolerance, Flowers and Flowers (2005) could do little more than point out the problems and failures of approaches ranging from conventional breeding to marker assisted selection to the use of transgenic plants. Munns (2005), advocated exploiting ‘candidate genes’ involved in salt uptake or transport, having osmotic or other protective functions, or that ‘make a plant grow more quickly in saline soil.’ At the same time, however, she noted that, ‘little has been revealed by gene expression studies so far, perhaps because the studies are not tissue-specific and because the treatments are often traumatic and unnatural’.
These assessments are correct and important, but as they have not been widely incorporated into actual research studies, they bear comment and extension. Overall, the two most critical factors influencing the outcome of stress studies are, arguably, growth of the plants and application of the stress. Because salinity is really only of interest in some ecological context, be it a natural or agricultural environment, if field studies are not possible, growth conditions should at least nominally reflect them (see also Hirayama and Shinozaki 2010). In general, this precludes using plants in Petri dishes at very low light and inclusion of sucrose as a carbon source, although as part of larger studies, such conditions may, potentially, have a place. In general, it also precludes growing plants in solutions with nutrient levels so high that they would present a major eutrophication problem were they to occur in nature, e.g. Hoagland solution, which, undiluted, has 15 mM nitrate and 0.5–1.0 mM phosphate, or 20–100 times the levels in most soils. Moreover, it requires consideration of the ratios of nutrient elements that might occur naturally. In seawater, the Na : K and Na : Ca ratios are 45 : 1 and the Ca : Mg ratio is 1 : 5. Studies of coastal halophytes should, arguably, be based on such a medium, not some NaCl supplemented version of Hoagland’s. In agricultural conditions, the Na : K ratio will depend on the source of the sodium. It would certainly be useful if experimental conditions would be chosen carefully and justified based on some field measurements.
I note that despite repeated criticism, use of shock treatments to plants with no previous stress experience is still the norm. This needs to stop. Munns (2005) provided an a propos list of what a realistic approach should include. It should: avoid large osmotic shocks; avoid measurements while plants are still recovering from the initial shock such as when cells are plasmolysed; avoid generating other nutrient imbalances or deficiencies, especially calcium, while applying the stress; measure physiological traits during treatments and, at harvest, determine dry weights rather than simply taking ‘representative’ photographs; and measure relative growth rate (RGR) rather than simply reporting final plant sizes. I would add that to make RGR interpretable, the determinations should include multiple measurements during the growth period, not just the first and last weights (e.g. Wickens and Cheeseman 1988; Cheeseman and Tankou 2005).
In this review, I have focussed primarily on two issues. The first is the misdirection which has characterised ‘salt tolerance’ research in the past few decades. The second is the identification of critical topics about which we know too little and which represent major gaps in the fundamental understanding the integration of activity, particularly in saline environments. How can these be moved forward?
Critical topics for further study
In Fig. 1, I identified seven broad aspects of plant development and metabolism which need to be integrated if an organism is to succeed in a saline environment. I feel obligated to suggest how to proceed with this effort. Each of the seven aspects requires two types of continued study. Hypothesis-driven studies that aggressively and rigorously challenge accepted conclusions and models are needed while at the same time extending the knowledge base for well-defined problems. It is of paramount importance, of course, that these hypotheses not be dependent on ambiguously defined terms (e.g. tolerance or toxicity).
There is also is a critical need for continued discovery-based research, targeting both principles of integration and functions of uncharacterised genes and their products. For example, in all genomes annotated to date, the number of unknown, putative protein and hypothetical protein encoding genes is ~50% of the total. These can be further divided into those conserved between species and those which are lineage specific (a.k.a. ‘orphan’ genes) (Emrich et al. 2007; Hanada et al. 2008; Khalturin et al. 2009; Oh et al. 2012). Hypothesis driven studies are needed to verify placement of genes in expression networks; discovery studies are needed for characterising the products of the unknown genes and their functions. The targets for these efforts should be selected based on, for example, RNAseq analyses of plants subjected to stress treatments appropriately applied.
Discovery studies are also needed with respect to known proteins. The LEAs, for example, effect stress tolerance without direct energy input, but their modes of action and in particular their associations with other proteins and complexes are poorly understood.
The characterisation of the cytosol of plant cells is a clear case of a critical problem needing input from both biophysics and protein chemistry. It will quite probably be impossible to solve using the complex tissues and vacuole-dominated cell structures of higher plants. For this, therefore, it makes more sense to return to the use of large celled algae where the cytosol can be readily isolated from vacuoles and characterised.
For all of these efforts, new approaches embracing systems biology are needed. These should minimise use of single gene transfers, employ knockout mutants wisely as part of whole system analyses, and use ‘phenotype’ as, at most, something indicating the presence of an interesting result (but not the result itself). We need to approach plants as complex systems built on redundant systems and interacting processes where no single gene alteration will bring about either catastrophic failure or incredible improvement. If we are going to use treatments that kill plants, we should analyse how the complex system fails. The RNAi approach to analysis of HSP90 functions (Sangster et al. 2004) is an a propos starting model for such efforts.
Towards a better understanding of salt ‘stress’ and ‘tolerance’
Having so uncharitably characterised the bulk of salt tolerance work as based on undefined terms and unquantifiable parameters, I conclude by addressing the question: can there be a useful exploration of ‘salt stress’ and is there any way to discuss ‘tolerance’ that goes beyond the simplistic phenological observations to which I have so vehemently objected? Briefly, the answer to the first questions is ‘yes, but it won’t be trivial’. To the second, the answer is ‘maybe.’
Eventually, the understanding of ‘salt stress’ and plant responses will depend on all aspects of a plant’s ‘physiology’, i.e. the sum of all cellular processes including biochemical and biophysical processes and transcriptome-, proteome- and metabolome complements, the epigenome, the small RNAs and as yet to be discovered regulatory systems as they are manifested at the organismal level.
To define stress, I will make an analogy to mechanical engineering where it refers to a deformational force applied to an object; the object’s response (deformation) is referred to as a strain. This is not new. I am not breaking new ground, but the distinctions are important and useful. In the context of environmental agents, a stress can be defined as any environmental perturbation. The associated strain is the physiological response of the plant, essentially a multidimensional space analogous to linear or angular deformation in material strength tests. There is no question that using microarray or RNAseq methods to document these spaces will provide an even more ‘bewildering array of data’ than the summary of physiological responses by Flowers et al. (1977) could envision. However, software for analysing and visualising these spaces is now readily available and continues to be developed and improved rapidly.
Technically, an environmental perturbation is a stress whether or not it results in a measurable strain, and a stress that produces a marked strain in one species or genotype may have little or no effect in another. Comparisons of the transcriptome responses of the salinity responses of A. thaliana and Thellungiella halophila (salsuginea) are good examples of this (e.g. Gong et al. 2005; Amtmann 2009). Nevertheless, it is clear that to be ‘interesting’, the stress must produce a change in, for example, the transcript complement or enzyme activation beyond that which would occur as part of normal diurnal or developmental patterns. As an aside, this also allows us to answer the question which arises frequently in informal discussion, ‘what is salt stress to a halophyte?’ Clearly, it is a manipulation of external salinity levels that produces a measurable strain, i.e. a persistent change in the transcript complement (or other -omic representation) of the plant. The minimal stress required to produce such a strain will normally be different in any two genotypes and certainly between any two species.
The method of application of the stress, especially its amplitude and rapidity, unquestionably affects the resultant strain. Shock treatments are easy to do and produce clear (in that they are visible) responses. However, in the context of understanding integrated activity or agronomic performance, dying plants are not very interesting. To get the most return for the effort and expense of an -omic level study, the strains need to be measured over a range of stress intensities, not just at the breaking point.
So, will there finally be a way to discuss ‘tolerance’ that goes beyond the simplistic phenological observations? Maybe. Between stresses that result in death and those which produce no measurable strain, we can expect, based on shock studies, a range of responses. With small stresses, we can expect minimal strains that relax to the pre-stress levels rapidly. With larger stresses, we can expect strains that relax to a new quasi-steady-state in which growth and development continue. Eventually, a stress may be large enough to induce strains that do not relax. When we have reached enlightenment, we will perhaps be able to select and agree on one point as that which defines ‘tolerance’.
Concluding remarks
In closing, I would like to issue a long overdue apology for completely ignoring, in preparing this review, some truly significant recent work pertinent to saline environments, work that needs separate attention and sets new important directions. I have ignored trees and perennials in general, despite the fact that they are the dominant organisms in significant and extensive saline environments. I have not discussed rhizomatous herbs and trees that can effectively move about and select their habitats (Salzman 1985; D’Hertefeldt et al. 2011), thus, substantially controlling their own resource environment. Despite the growing body of research on the topic, I have ignored the influence of rhizosphere microbes, including bacteria, mycorrhizal fungi and N-fixing symbionts (see Aroca and Ruiz-Lozano 2009; Miransari 2010; Porcel et al. 2012). Finally, I have discussed saline environments as though they existed in generic, ubiquitously comparable ways, failing to note the importance of chlorides and sulfates, of sodic and alkaline soils of the balance of other essential and potentially deleterious ions, of the limitations imposed by diffusion of ions through soil and of all the other factors associated with ecosystems ranging from tropical coastal mangroves, to subtropical and temperate salinised agroecosystems and to north temperate/boreal over-grazed semideserts.
Acknowledgements
I would like to gratefully acknowledge critical readings of early versions of this review by Carol Augspurger (University of Illinois), for their helpful discussions, Maheshi Dassanayake, Dong-Ha Oh, Hyewon Hong and Hans Bohnert (University of Illinois and Gyeongsang National University, Jinju, Korea).
References
Alcázar R, Bitrián M, Bartels D, Koncz C, Altabella T, Tiburcio AF (2011) Polyamine metabolic canalization in response to drought stress in Arabidopsis and the resurrection plant Craterostigma plantagineum. Plant Signaling & Behavior 6, 243–250.| Polyamine metabolic canalization in response to drought stress in Arabidopsis and the resurrection plant Craterostigma plantagineum.Crossref | GoogleScholarGoogle Scholar |
Ali AJ, Xu JL, Ismail AM, Fu BY, Vijaykumar CHM, Gao YM, Domingo J, Maghirang R, Yu SB, Gregorio G, Yanaghihara S, Cohen M, Carmen B, Mackill D, Li ZK (2006) Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Research 97, 66–76.
| Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program.Crossref | GoogleScholarGoogle Scholar |
Allen RD (1995) Dissection of oxidative stress tolerance using transgenic plants. Plant Physiology 107, 1049–1054.
Amtmann A (2009) Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Molecular Plant 2, 3–12.
| Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXovV2msbk%3D&md5=1ab76ac13e1dad8d7821e44289ab1f50CAS |
Amtmann A, Hammond JP, Armengaud P, White PJ (2006) Nutrient sensing and signalling in plants: potassium and phosphorus. In ‘Advances in botanical research’. (Ed. JA Callow) pp. 209–257. (Academic Press: New York)
Apse MP, Blumwald E (2002) Engineering salt tolerance in plants. Current Opinion in Biotechnology 13, 146–150.
| Engineering salt tolerance in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xis1CltL8%3D&md5=51a8027c374160a13b1a4715c0f5dcf9CAS |
Aroca R, Ruiz-Lozano JM (2009) Induction of plant tolerance to semi-arid environments by beneficial soil microorganisms – a review. In ‘Climate change, intercropping, pest control and beneficial microorganisms’. (Ed. E Lichtfouse) pp. 121–135. (Springer Science and Business Media, B.V.: Dordrecht, The Netherlands)
Aroca R, Porcel R, Ruiz-Lozano JM (2012) Regulation of root water uptake under abiotic stress conditions. Journal of Experimental Botany 63, 43–57.
| Regulation of root water uptake under abiotic stress conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs1yms7rP&md5=30009a5f7ee07066a862aeec42f2af9eCAS |
Asada K (1994) Mechanism of radical scavenging in chloroplasts under light stress. In ‘Photoinhibition of photosynthesis – from molecular mechanisms to the field’. (Eds NR Baker, JR Bowyer) pp. 129–142. (Bios Scientific Publishers: Oxford)
Attia H, Karray N, Rabhi M, Lachaal M (2008) Salt-imposed restrictions on the uptake of macroelements by roots of Arabidopsis thaliana. Acta Physiologiae Plantarum 30, 723–727.
| Salt-imposed restrictions on the uptake of macroelements by roots of Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXitVWmsrs%3D&md5=521dd34110e0048fd749773208dbf00aCAS |
Bae EK, Lee H, Lee JS, Noh EW (2011) Drought, salt and wounding stress induce the expression of the plasma membrane intrinsic protein 1 gene in poplar (Populus alba × P. tremula var. glandulosa). Gene 483, 43–48.
| Drought, salt and wounding stress induce the expression of the plasma membrane intrinsic protein 1 gene in poplar (Populus alba × P. tremula var. glandulosa).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXos1SlsLs%3D&md5=69cb7caf0db095926d7852f289ef4a7aCAS |
Batchelder EM, Yarar D (2010) Differential requirements for clathrin-dependent endocytosis at sites of sell-substrate adhesion. Molecular Biology of the Cell 21, 3070–3079.
| Differential requirements for clathrin-dependent endocytosis at sites of sell-substrate adhesion.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtlGlt7jP&md5=c3f4a089244f9f97c4cde828f00539c5CAS |
Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiology 148, 6–24.
| The enigmatic LEA proteins and other hydrophilins.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFKnurnL&md5=af6f3e9610ae64965718f965092613d0CAS |
Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, Vitek O, Salt DE (2010) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLOS Genetics 6, e1001193
| A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1.Crossref | GoogleScholarGoogle Scholar |
Bazihizina N, Colmer TD, Barrett-Lennard EG (2009) Response to non-uniform salinity in the root zone of the halophyte Atriplex nummularia: growth, photosynthesis, water relations and tissue ion concentrations. Annals of Botany 104, 737–745.
| Response to non-uniform salinity in the root zone of the halophyte Atriplex nummularia: growth, photosynthesis, water relations and tissue ion concentrations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtF2ktbjE&md5=25547b7b68aa56a3a31b41f39a3dee47CAS |
Bazihizina N, Barrett-Lennard EG, Colmer TD (2012) Plant growth and physiology under heterogeneous salinity. Plant and Soil 354, 1–19.
| Plant growth and physiology under heterogeneous salinity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XlvFWgu7w%3D&md5=ef568629c88200f6234b93cddce048c5CAS |
Behal RH, Buxton DB, Robertson JG, Olson MS (1993) Regulation of the pyruvate-dehydrogenase multienzyme complex. Annual Review of Nutrition 13, 497–520.
| Regulation of the pyruvate-dehydrogenase multienzyme complex.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXlslKgsrY%3D&md5=11754d0e6867323305d764c48139e3fdCAS |
Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, Gosti F, Simonneau T, Essah PA, Tester M, Véry AA, Sentenac H, Casse F (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. The EMBO Journal 22, 2004–2014.
| Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjsVKmu70%3D&md5=c5f9279ade6122015f7d8f9e7b4814b7CAS |
Bertrand G, Perietzeanu J (1927) Sur la présence du sodium chez les plantes. Compte Rendus de la Séance 184, 645–649.
Book AJ, Gladman NP, Lee S-S, Scalf M, Smith LM, Vierstra RD (2010) Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. The Journal of Biological Chemistry 285, 25 554–25 569.
| Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXpslKjtb8%3D&md5=3a4b2448d75774adadf4dc8ec484b92eCAS |
Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C (2005) Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiology 139, 790–805.
| Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtFCgsb7I&md5=a33150b1e4cf10670e11f666fc1bb783CAS |
Britto D, Kronzucker HJ (2009) Ussing’s conundrum and the search for transport mechanisms in plants. New Phytologist 183, 243–246.
| Ussing’s conundrum and the search for transport mechanisms in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXptlSjsrs%3D&md5=f1f73828c83954bf5d9be33539ad231fCAS |
Carden DE, Walker DJ, Flowers TJ, Miller AJ (2003) Single-cell measurements of the contributions of cytosolic Na+ and K+ to salt tolerance. Plant Physiology 131, 676–683.
| Single-cell measurements of the contributions of cytosolic Na+ and K+ to salt tolerance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXhtlyjs70%3D&md5=765b844d93057ca5162d3b32f491bea4CAS |
Chao DY, Luo YH, Shi M, Luo D, Lin HX (2005) Salt-responsive genes in rice revealed by cDNA microarray analysis. Cell Research 15, 796–810.
| Salt-responsive genes in rice revealed by cDNA microarray analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtlSgt7zK&md5=4754ccc3cb621542e5d80afc7c6411caCAS |
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Annals of Botany 103, 551–560.
| Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXktVGnu7s%3D&md5=c0a3d54246b0668e76fa3410ea5d90a2CAS |
Chaves MM, Costa JM, Saibo NJM (2011) Recent advances in photosynthesis under drought and salinity. In ‘Plant responses to drought and salinity stress: developments in a post-genomic era’. (Ed. I Turkan) pp. 49–104. (Academic Press: New York)
Cheeseman JM (1988) Mechanisms of salinity tolerance in plants. Plant Physiology 87, 547–550.
| Mechanisms of salinity tolerance in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1cXltVOlt70%3D&md5=99b04d0af46e7d767cb00e6e9e7bbee3CAS |
Cheeseman JM (2006) Hydrogen peroxide concentrations in leaves under natural conditions. Journal of Experimental Botany 57, 2435–2444.
| Hydrogen peroxide concentrations in leaves under natural conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XnvF2lsrY%3D&md5=8934d9e8a846a987fb7b9b3d98586368CAS |
Cheeseman J (2007) Hydrogen peroxide and plant stress: a challenging relationship. Plant Stress 1, 4–15.
Cheeseman J (2009) Seasonal patterns of leaf H2O2 content: reflections of leaf phenology, or environmental stress? Functional Plant Biology 36, 721–731.
| Seasonal patterns of leaf H2O2 content: reflections of leaf phenology, or environmental stress?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXovVert7s%3D&md5=b6923f9aa6eace731c73e53bfa578723CAS |
Cheeseman JM, Lexa M (1996) Gas exchange: models and measurements. In ‘Photosynthesis and the environment’. (Ed. NR Baker) pp. 223–240. (Kluwer Academic Publishers: Dordrecht, The Netherlands)
Cheeseman J, Lovelock C (2004) Photosynthetic characteristics of dwarf and fringe Rhizophora mangle L. in a Belizean mangrove. Plant, Cell & Environment 27, 769–780.
| Photosynthetic characteristics of dwarf and fringe Rhizophora mangle L. in a Belizean mangrove.Crossref | GoogleScholarGoogle Scholar |
Cheeseman JM, Tankou S (2005) Nitrate reductase and growth of Arabidopsis thaliana in solution culture. Plant and Soil 266, 143–152.
| Nitrate reductase and growth of Arabidopsis thaliana in solution culture.Crossref | GoogleScholarGoogle Scholar |
Cheeseman JM, Wickens LK (1986) Control of Na+ and K+ transport in Spergularia marina iii. Relationship between ion uptake and growth at moderate salinity. Physiologia Plantarum 67, 15–22.
| Control of Na+ and K+ transport in Spergularia marina iii. Relationship between ion uptake and growth at moderate salinity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL28Xkt1WqtL8%3D&md5=a51f004f6f75a1caf54819ca82ed90d1CAS |
Cheeseman JM, Bloebaum P, Wickens LK (1985) Short term 22Na+ and 42K+ uptake in intact, mid-vegetative Spergularia marina plants. Physiologia Plantarum 65, 460–466.
| Short term 22Na+ and 42K+ uptake in intact, mid-vegetative Spergularia marina plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL28XosFCqtw%3D%3D&md5=18ced08fc9adf25bcce1749e867a5c0dCAS |
Cheeseman JM, Clough BF, Carter DR, Lovelock CE, Eong OJ, Sim RG (1991) The analysis of photosynthetic performance in leaves under field conditions: a case study using Bruguiera mangroves. Photosynthesis Research 29, 11–22.
Cheeseman JM, Herendeen LB, Cheeseman AT, Clough BF (1997) Photosynthesis and photoprotection in mangroves under field conditions. Plant, Cell & Environment 20, 579–588.
| Photosynthesis and photoprotection in mangroves under field conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXjsFamurg%3D&md5=5247028431441d8b38448585d803a1b9CAS |
Clarkson DT, Hanson JB (1980) The mineral nutrition of higher plants. Annual Review of Plant Physiology 31, 239–298.
| The mineral nutrition of higher plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3cXks12msL0%3D&md5=ed8e44a6a6292f4385c76adcb62d896aCAS |
Collander R (1941) Selective absorption of cations by higher plants. Plant Physiology 16, 691–720.
| Selective absorption of cations by higher plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaH3sXmvVWr&md5=01eb3139ba2ca3f8ce083a1247a9e428CAS |
Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M (2012) A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 7, e39865
| A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtVKrsrvM&md5=69ba86efd120a894346aa07d3f4e9ce7CAS |
Cram W, Torr P, Rose D (2002) Salt allocation during leaf development and leaf fall in mangroves. Trees – Structure and Function 16, 112–119.
Csermely P, Söti C, Blatch G (2007) Chaperones as parts of cellular networks. In ‘Molecular aspects of the stress response: chaperones, membranes and networks’. (Eds P Csermely, L Vígh) pp. 55–63. (Landes Bioscience and Springer Science+Business Media: New York, NY)
D’Hertefeldt T, Falkengren-Grerup U, Jonsdottir IS (2011) Responses to mineral nutrient availability and heterogeneity in physiologically integrated sedges from contrasting habitats. Plant Biology 13, 483–492.
| Responses to mineral nutrient availability and heterogeneity in physiologically integrated sedges from contrasting habitats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXlvFyns7Y%3D&md5=7c3e74e68adfdfcb89c5ed401430caaaCAS |
Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun D-J, Bressan RA, Zhu J-K, Bohnert HJ, Cheeseman JM (2011) The genome of the extremophile crucifer Thellungiella parvula. Nature Genetics 43, 913–918.
| The genome of the extremophile crucifer Thellungiella parvula.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXpvVOltLo%3D&md5=4a9abf9a293e9aca54fe119071b17588CAS |
Davenport RJ, Munoz-Mayor A, Jha D, Essah PA, Rus A, Tester M (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant, Cell & Environment 30, 497–507.
| The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXksVemu7Y%3D&md5=eb6fd1dc37317581f21af0cde6b91ba0CAS |
De Boer AH, Volkov V (2003) Logistics of water and salt transport through the plant: structure and functioning of the xylem. Plant, Cell & Environment 26, 87–101.
| Logistics of water and salt transport through the plant: structure and functioning of the xylem.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXhtlKrtLk%3D&md5=af33c1a02a68e408fe74e192ac07d827CAS |
Dettmer J, Elo A, Helariutta Y (2009) Hormone interactions during vascular development. Plant Molecular Biology 69, 347–360.
| Hormone interactions during vascular development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhvVOis7k%3D&md5=07ee7fef0e227fd6fbb9cf53d9c6dabcCAS |
Emrich SJ, Barbazuk WB, Li L, Schnable PS (2007) Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Research 17, 69–73.
| Gene discovery and annotation using LCM-454 transcriptome sequencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXms1Khug%3D%3D&md5=49d2bd9f2a172008bd0606f25acf7716CAS |
Essah PA, Davenport R, Tester M (2003) Sodium influx and accumulation in Arabidopsis. Plant Physiology 133, 307–318.
| Sodium influx and accumulation in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXntlaiur8%3D&md5=d50c579b0060138d3ddeb00baf2082b8CAS |
Everard JD, Gucci R, Kann SC, Flore JA, Loescher WH (1994) Gas exchange and carbon partitioning in the leaves of celery (Apium graveolens L) at various levels of root zone salinity. Plant Physiology 106, 281–292.
Faiyue B, Al-Azzawi MJ, Flowers TJ (2010) The role of lateral roots in bypass flow in rice (Oryza sativa L.). Plant, Cell & Environment 33, 702–716.
Flexas J, Bota J, Loreto F, Cornic G, Sharkey TD (2004) Diffusive and metabolic limitations to photosynthesis under drought and salinity in C3 plants. Plant Biology 6, 269–279.
| Diffusive and metabolic limitations to photosynthesis under drought and salinity in C3 plants.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD2c3ksVOlug%3D%3D&md5=65389de98cf76891620aadf2aa3ba8f8CAS |
Flowers T (1972) Salt tolerance in Suaeda maritima (L.) Dum. Journal of Experimental Botany 23, 310–321.
| Salt tolerance in Suaeda maritima (L.) Dum.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE38XksVGmurY%3D&md5=3f99e48c70b6c547ba17ac7554631be6CAS |
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytologist 179, 945–963.
| Salinity tolerance in halophytes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFWqur%2FE&md5=06dcabcbf1411c34a35d7aed78f04afbCAS |
Flowers T, Flowers S (2005) Why does salinity pose such a difficult problem for plant breeders. Agricultural Water Management 78, 15–24.
| Why does salinity pose such a difficult problem for plant breeders.Crossref | GoogleScholarGoogle Scholar |
Flowers TJ, Läuchli A (1983) Sodium versus potassium: substitution and compartmentation. In ‘Inorganic plant nutrition’. (Eds A Läuchli, A Pirson) pp. 651–681. (Springer-Verlag: Berlin)
Flowers T, Ward M, Hall J (1976) Salt tolerance in the halophyte Suaeda maritima: some properties of malate dehydrogenase. Philosophical Transactions of the Royal Society of London, Series B 273, 523–540.
| Salt tolerance in the halophyte Suaeda maritima: some properties of malate dehydrogenase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE28XktValu78%3D&md5=c2c84095df9e5ad0c719f0a5ed9ef0baCAS |
Flowers TJ, Troke PF, Yeo AR (1977) The mechanism of salt tolerance in halophytes. Annual Review of Plant Physiology 28, 89–121.
| The mechanism of salt tolerance in halophytes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE2sXksFSisb8%3D&md5=ab0a16957d0ffbee896436c71cd4eb84CAS |
Flowers TJ, Galal HK, Bromham L (2010) Evolution of halophytes: multiple origins of salt tolerance in land plants. Functional Plant Biology 37, 604–612.
| Evolution of halophytes: multiple origins of salt tolerance in land plants.Crossref | GoogleScholarGoogle Scholar |
Forde BG (2009) Is it good noise? The role of developmental instability in the shaping of a root system. Journal of Experimental Botany 60, 3989–4002.
| Is it good noise? The role of developmental instability in the shaping of a root system.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXht1WqtLbP&md5=4e2ebecc6ace9eca095322d27ed9cc7cCAS |
Friesen ML, Cordeiro MA, Penmetsa RV, Badri M, Huguet T, Aouani ME, Cook DR, Nuzhdin SV (2010) Population genomic analysis of Tunisian Medicago truncatula reveals candidates for local adaptation. The Plant Journal 63, 623–635.
| Population genomic analysis of Tunisian Medicago truncatula reveals candidates for local adaptation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtFegtrbE&md5=accec68c18314a22602d2c598595fa9bCAS |
Galinha C, Bilsborough G, Tsiantis M (2009) Hormonal input in plant meristems: a balancing act. Seminars in Cell & Developmental Biology 20, 1149–1156.
| Hormonal input in plant meristems: a balancing act.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsVGntL3J&md5=421e30f83aec5af614074ea84bb04ac6CAS |
Genc Y, McDonald GK, Tester M (2007) Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant, Cell & Environment 30, 1486–1498.
| Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXht1yhsrbE&md5=4d23774f14c29efebba1e085d413389eCAS |
Golldack D, Luking I, Yang O (2011) Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Reports 30, 1383–1391.
| Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXovVWgtrY%3D&md5=8ff7a0dcf1e7bea673dd1733f63ac56bCAS |
Gong QQ, Li PH, Ma SS, Rupassara SI, Bohnert HJ (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. The Plant Journal 44, 826–839.
| Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtlWltrbN&md5=620030b0bcdb8cf64087da8a830c4207CAS |
Graham JWA, Williams TCR, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ (2007) Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. The Plant Cell 19, 3723–3738.
| Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXns1eltQ%3D%3D&md5=10d09344ac6e076ff0169082a75a272fCAS |
Greenway H, Munns R (1980) Mechanisms of salt tolerance in nonhalophytes. Annual Review of Plant Physiology 31, 149–190.
| Mechanisms of salt tolerance in nonhalophytes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3cXksVWntb4%3D&md5=03b2e3789f9318b2329214779e4586beCAS |
Greenway H, Osmond C (1972) Salt responses of enzymes from species differing in salt tolerance. Plant Physiology 49, 256–259.
| Salt responses of enzymes from species differing in salt tolerance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE38Xps1amtg%3D%3D&md5=7b303cda1e3881933311dfa1b85f3f8eCAS |
Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology 132, 578–596.
| Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXkslersbo%3D&md5=a3fb747518533b8c8120e0e77c3dd821CAS |
Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu S-H (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiology 148, 993–1003.
| Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXht1GmtLjN&md5=e511d52b3552dd536b22438871437657CAS |
Haslam E (1986) Secondary metabolism – fact and fiction. Natural Product Reports 3, 217–249.
| Secondary metabolism – fact and fiction.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL28XmtVCrs7w%3D&md5=cd86d312b87598f699b14645272fd170CAS |
Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in higher molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiology 149, 1541–1559.
| Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in higher molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXlsFCiurc%3D&md5=7f340cf2a0318b82f1ab349543395cf7CAS |
Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. The Plant Journal 61, 1041–1052.
| Research on plant abiotic stress responses in the post-genome era: past, present and future.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXkvFKntL4%3D&md5=9c1512a14114003ab0d2734c0f6c4050CAS |
Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung H-Y, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. The EMBO Journal 26, 3003–3014.
| Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXms1WntLg%3D&md5=adf87843dd9889971383ce66881e93d4CAS |
Horie T, Hauser F, Schroeder JI (2009) HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends in Plant Science 14, 660–668.
| HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsV2mtr7J&md5=bd10537d971a3724f00ab6490c5146ffCAS |
Iglesias MJ, Terrile MC, Bartoli CG, D’Ippolito S, Casalongue CA (2010) Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Molecular Biology 74, 215–222.
| Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtFGhurbP&md5=5dd553aa3273ed7173c6ee4b62658812CAS |
Jacoby B (1979) Sodium recircumation and loss from Phaseolus vulgaris L. Annals of Botany 43, 741–744.
Jennings DH (1976) The effects of sodium chloride on higher plants. Biological Reviews of the Cambridge Philosophical Society 51, 453–486.
| The effects of sodium chloride on higher plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE2sXhtF2kurw%3D&md5=22ee17c76ce50faade3dca90e1eb9a18CAS |
Kandil FE, Grace MH, Seigler DS, Cheeseman JM (2004) Polyphenolics in Rhizophora mangle L. leaves and their changes during leaf development and senescence. Trees 18, 518–528.
| Polyphenolics in Rhizophora mangle L. leaves and their changes during leaf development and senescence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXms1Kqu78%3D&md5=f94cdddc3a53c33d46816e4eeab7e666CAS |
Karley AJ, Leigh RA, Sanders D (2000) Where do all the ions go? The cellular basis of differential ion accumulation in leaf cells. Trends in Plant Science 5, 465–470.
| Where do all the ions go? The cellular basis of differential ion accumulation in leaf cells.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3M%2FmtFSmsQ%3D%3D&md5=2b4cb3aec579f02a617bd1d9c022d34aCAS |
Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG (2009) More than just orphans: are taxonomically-restricted genes important in evolution? Trends in Genetics 25, 404–413.
| More than just orphans: are taxonomically-restricted genes important in evolution?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtV2rsLnI&md5=73f9364a480d1d61ede2ad1e783c7846CAS |
Kim W-Y, Ali Z, Park HJ, Park SJ, Cha J-Y, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z (2013) Release of SOS2 kinase from sequestration with GIGANTIA determines salt tolerance in Arabidopsis. Nature Communications 4,
| Release of SOS2 kinase from sequestration with GIGANTIA determines salt tolerance in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | in press
Kong XQ, Luo Z, Dong HH, Eneji AE, Li WJ (2012) Effects of non-uniform root zone salinity on water use, Na+ recirculation, and Na+ and H+ flux in cotton. Journal of Experimental Botany 63, 2105–2116.
| Effects of non-uniform root zone salinity on water use, Na+ recirculation, and Na+ and H+ flux in cotton.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjslehsLk%3D&md5=8660da5ca953aef5c12c61378161c6c2CAS |
Kronzucker HJ, Britto DT (2011) Sodium transport in plants: a critical review. New Phytologist 189, 54–81.
| Sodium transport in plants: a critical review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXltlGhug%3D%3D&md5=4ead47138c07cc9af919c11d4c2fd438CAS |
Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM (2010) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biology 11, R123
| Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXktlSjsA%3D%3D&md5=bc6c990af81d4430f634eb5f4dd58332CAS |
Lambers H, Chapin FS, III, Pons TL (2008) ‘Plant physiological ecology.’ (Springer: New York)
Läuchli A, James R, Huang C, McCully M, Munns R (2008) Cell-specific localization of Na+ in roots of durum wheat and possible control points for salt exclusion. Plant, Cell & Environment 31, 1565–1574.
| Cell-specific localization of Na+ in roots of durum wheat and possible control points for salt exclusion.Crossref | GoogleScholarGoogle Scholar |
Lazof D, Cheeseman JM (1986) Sodium transport and compartmentation in Spergularia marina: partial characterization of a functional symplasm. Plant Physiology 81, 742–747.
| Sodium transport and compartmentation in Spergularia marina: partial characterization of a functional symplasm.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL28XkslKju7g%3D&md5=6b1f76d94d4a382ba4354624e0619996CAS |
Li XJ, Wang XH, Yang Y, Li RL, He QH, Fang XH, Luu DT, Maurel C, Lin JX (2011) Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. The Plant Cell 23, 3780–3797.
| Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs1eku7rF&md5=b86eca268af8e5869545d94f3ccd680fCAS |
Liu F, Makhmoudova A, Lee EA, Wait R, Emes MJ, Tetlow IJ (2009) The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. Journal of Experimental Botany 60, 4423–4440.
| The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtlGitL7E&md5=e964bef186c857f1ed362797fa323a1eCAS |
López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology 6, 280–287.
| The role of nutrient availability in regulating root architecture.Crossref | GoogleScholarGoogle Scholar |
Lu M, Sautin Y, Holliday L, Gluck S (2004) The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. The Journal of Biological Chemistry 279, 8732–8739.
| The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhs1OntL8%3D&md5=02ce996c8437567cea8033fe9fc82330CAS |
Lundegårdh H (1929) ‘Die quantitative Spektralanalyse der Elemente und ihre Anwendung auf biologische, agrikulturchemische ind mineralogische Aufgaben.’ (Verlag von Gustav Fischeer: Jena)
Luu DT, Martiniere A, Sorieul M, Runions J, Maurel C (2012) Fluorescence recovery after photobleaching reveals high cycling dynamics of plasma membrane aquaporins in Arabidopsis roots under salt stress. The Plant Journal 69, 894–905.
| Fluorescence recovery after photobleaching reveals high cycling dynamics of plasma membrane aquaporins in Arabidopsis roots under salt stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XktFWjtrk%3D&md5=3840ca6096325ec69dc1bda4f2005adeCAS |
Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biology 8, R49
| Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression.Crossref | GoogleScholarGoogle Scholar |
Ma PD, Liu JY (2012) Isolation and characterization of a novel plasma membrane intrinsic protein gene, LcPIP1, in Leymus chinensis that enhances salt stress tolerance in Saccharomyces cerevisiae. Applied Biochemistry and Biotechnology 166, 479–485.
| Isolation and characterization of a novel plasma membrane intrinsic protein gene, LcPIP1, in Leymus chinensis that enhances salt stress tolerance in Saccharomyces cerevisiae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XosFOjsw%3D%3D&md5=6ef13d48e67fb3edd936319546080fdcCAS |
Maathuis FJM (2007) Monovalent cation transporters; establishing a link between bioinformatics and physiology. Plant and Soil 301, 1–15.
| Monovalent cation transporters; establishing a link between bioinformatics and physiology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXosVaitg%3D%3D&md5=fa4708e3de75db613c1ac55bc9d93c7bCAS |
Malagoli P, Britto DT, Schulze LM, Kronzucker HJ (2008) Futile Na+ cycling at the root plasma membrane in rice (Oryza sativa L.): kinetics, energetics, and relationship to salinity tolerance. Journal of Experimental Botany 59, 4109–4117.
| Futile Na+ cycling at the root plasma membrane in rice (Oryza sativa L.): kinetics, energetics, and relationship to salinity tolerance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsVCgt73O&md5=88a6a2ad1bb21a9f7670a3a5c98f6f2eCAS |
Maurel C, Verdoucq L, Luu D, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Annual Review of Plant Biology 59, 595–624.
| Plant aquaporins: membrane channels with multiple integrated functions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXntFaqtr4%3D&md5=a8c5b1244d3e8d4c5fbe3e494e6aee32CAS |
Maurel C, Santoni V, Luu D, Wudick M, Verdoucq L (2009) The cellular dynamics of plant aquaporin expression and functions. Current Opinion in Plant Biology 12, 690–698.
| The cellular dynamics of plant aquaporin expression and functions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsV2murnM&md5=147268ae59e8340e9213e27737a4e943CAS |
McNeil DL, Atkins CA, Pate JS (1979) Uptake and utilization of xylem-borne amino compounds by shoot organs of a legume. Plant Physiology 63, 1076–1081.
| Uptake and utilization of xylem-borne amino compounds by shoot organs of a legume.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE1MXkslWkt7g%3D&md5=7165bb36458226aa3e49c3abda93a1eeCAS |
Messedi D, Labidi N, Grignon C, Abdelly C (2004) Limits imposed by salt to the growth of the halophyte Sesuvium portulacastrum. Journal of Plant Nutrition and Soil Science 167, 720–725.
| Limits imposed by salt to the growth of the halophyte Sesuvium portulacastrum.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhsVer&md5=7a51c224bb8e6a4676c642f43a346c96CAS |
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species, homeostasis and signalling during drought and salinity stresses. Plant, Cell & Environment 33, 453–467.
| Reactive oxygen species, homeostasis and signalling during drought and salinity stresses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXltV2hur8%3D&md5=207df450b712ba15e4b185abc7d07028CAS |
Miransari M (2010) Contribution of arbuscular mycorrhizal symbiosis to plant growth under different types of soil stress. Plant Biology 12, 563–569.
Moller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. The Plant Cell 21, 2163–2178.
| Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytologist 167, 645–663.
| Genes and salt tolerance: bringing them together.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVGisbfP&md5=a2d23c6d5faa06931cb8a05ae3e289bcCAS |
Munns R (2011) Plant adaptations to salt and water stress: differences and commonalities. In ‘Plant responses to drought and salinity stress: developments in a post-genomic era’. (Ed. I Turkan) pp. 1–32. (Academic Press: New York)
Munns R, James R (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant and Soil 253, 201–218.
| Screening methods for salinity tolerance: a case study with tetraploid wheat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXltVemsbc%3D&md5=aa01ce01ce313fb5da5c0e864a48405eCAS |
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681.
| Mechanisms of salinity tolerance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXntFaqtrw%3D&md5=0ce1f7aad22c73fffb329d4603f0e836CAS |
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnology 30, 360–364.
Nayal M, DiCera E (1996) Valence screening of water in protein crystals reveals potential Na+ binding sites. Journal of Molecular Biology 256, 228–234.
| Valence screening of water in protein crystals reveals potential Na+ binding sites.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xht1Sgu70%3D&md5=c7f748d2e263a5f951b37a1bf47ea5caCAS |
Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer CH (2002) Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration. Annals of Botany 89, 841–850.
| Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XlsVeitLs%3D&md5=501ad82787ede534efdaf872c9df1089CAS |
Oh D-H, Dassanayake M, Haas JS, Kropornika A, Wright C, Paino d’Urzo M, Hong H, Ali S, Hernandez A, Lambert GM, et al (2010) Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiology 154, 1040–1052.
| Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsV2nsbfP&md5=87279db9296b926d0fb9d036cc19ccbeCAS |
Oh D, Dassanayake M, Bohnert H, Cheeseman J (2012) Life at the extreme: lessons from the genome. Genome Biology 13, 241–249.
| Life at the extreme: lessons from the genome.Crossref | GoogleScholarGoogle Scholar |
Olinares PDB, Ponnala L, van Wijk KJ (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Molecular & Cellular Proteomics 9, 1594–1615.
| Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtlGisbjF&md5=70e5323d06dabda8af15a297809baf69CAS |
Page MJ, Di Cera E (2006) Role of Na+ and K+ in enzyme function. Physiological Reviews 86, 1049–1092.
| Role of Na+ and K+ in enzyme function.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1SgtL%2FK&md5=c5177df7d8997b7647f6417d11dd29e6CAS |
Pardo JM, Quintero FJ (2002) Plants and sodium ions: keeping company with the enemy. Genome Biology 3, reviews1017.1–reviews1017.4.
Park J, Kim YY, Martinoia E, Lee Y (2008) Long-distance transporters of inorganic nutrients in plants. Journal of Plant Biology 51, 240–247.
| Long-distance transporters of inorganic nutrients in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtVajtL7P&md5=4256bc929d7fc22c75318b2f35893228CAS |
Pearse I, Heath KD, Cheeseman JM (2005) A partial characterization of peroxidase in Rhizophora mangle. Plant, Cell & Environment 28, 612–622.
| A partial characterization of peroxidase in Rhizophora mangle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXkslKlu7g%3D&md5=c5d9a43dcf76667fb4c183d35e46886fCAS |
Perham RN (2000) Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annual Review of Biochemistry 69, 961–1004.
| Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXnt1aju74%3D&md5=9978602fd0e47f7125574e23966e40c2CAS |
Plett DC, Møller IS (2010) Na+ transport in glycophytic plants: what we know and would like to know. Plant, Cell & Environment 33, 612–626.
| Na+ transport in glycophytic plants: what we know and would like to know.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXltV2hu7w%3D&md5=62d8291a79227b8a4be4b4b3ef8f1d5fCAS |
Porcel R, Aroca R, Ruiz-Lozano JM (2012) Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agronomy for Sustainable Development 32, 181–200.
| Salinity stress alleviation using arbuscular mycorrhizal fungi. A review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsFSitLk%3D&md5=bc20cf28a96c86f52c9f21aa8e126535CAS |
Rae L, Lao NT, Kavanagh TA (2011) Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta 234, 429–444.
| Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtVyrtrfJ&md5=e799b3909d0c752bf2724fbaea3cad37CAS |
Rodríguez-Gamir J, Ancillo G, Legaz F, Primo-Millo E, Forner-Giner MA (2012) Influence of salinity on PIP gene expression in citrus roots and its relationship with root hydraulic conductance, transpiration and chloride exclusion from leaves. Environmental and Experimental Botany 78, 163–166.
| Influence of salinity on PIP gene expression in citrus roots and its relationship with root hydraulic conductance, transpiration and chloride exclusion from leaves.Crossref | GoogleScholarGoogle Scholar |
Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proceedings of the National Academy of Sciences of the United States of America 108, 18 524–18 529.
| Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsFymu7bO&md5=843f02c775dcf9b6677004a8c050c0fdCAS |
Salzman A (1985) Habitat selection in a clonal plant. Science 228, 603–604.
| Habitat selection in a clonal plant.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL2M7mvVaqtQ%3D%3D&md5=9422b3f9513fe1aa1f743f93771b13acCAS |
Sangster TA, Lindquist S, Queitsch C (2004) Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. BioEssays 26, 348–362.
| Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjs1Oqsbs%3D&md5=9ebc14d772f04f450952e57e2c0ff06cCAS |
Sangster T, Salathia N, Lee HN, Watanabe E, Schellenberg K, Morneau K, Wang H, Undurraga S, Queitsch C, Lindquist S (2008a) HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 105, 2969
| HSP90-buffered genetic variation is common in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXjtVSisbs%3D&md5=bebc03579ad70c63448f5bce96c9e180CAS |
Sangster TA, Salathia N, Undurraga S, Milo R, Schellenberg K, Lindquist S, Queitsch C (2008b) HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proceedings of the National Academy of Sciences of the United States of America 105, 2963–2968.
| HSP90 affects the expression of genetic variation and developmental stability in quantitative traits.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXjtVSisbo%3D&md5=d659fe9bca7d466361c6d45bbe27c936CAS |
Schachtman DP, Shin R (2007) Nutrient sensing and signalling: NPKS. Annual Review of Plant Biology 58, 47–69.
| Nutrient sensing and signalling: NPKS.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXnsVahs74%3D&md5=34a6176ea9d542f49a99599f8908820bCAS |
Scholander P (1968) How mangroves desalinate seawater. Physiologia Plantarum 21, 251–261.
| How mangroves desalinate seawater.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaF1cXhtVamtb8%3D&md5=941301452f162e72263687324d964f9fCAS |
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal 31, 279–292.
| Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XntFelsLg%3D&md5=b473fbe86599d2919ad8c079b2e8c638CAS |
Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiologia Plantarum 133, 651–669.
| Potassium transport and plant salt tolerance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXps1Oit70%3D&md5=b00298e506b7189fc2da0a5277d6448aCAS |
Silvertown J, Poulton P, Johston E, Edwards G, Heard M, Biss PM (2006) The Park Grass Experiment 1856–2006: its contribution to ecology. Journal of Ecology 94, 801–814.
| The Park Grass Experiment 1856–2006: its contribution to ecology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XnsFWjsrc%3D&md5=be631ee1046f282f084038670bec3cb0CAS |
Spitzer J, Poolman B (2005) Electrochemical structure of the crowded cytoplasm. Trends in Biochemical Sciences 30, 536–541.
| Electrochemical structure of the crowded cytoplasm.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVKjsL7K&md5=dbdcf2683cc7c2c568aeacc83c335e61CAS |
Spitzer J, Poolman B (2009) The role of biomacromolecular crowding, ionic strength, and physiochemical gradients in the complexities of life’s emergence. Microbiology and Molecular Biology Reviews 73, 371–388.
| The role of biomacromolecular crowding, ionic strength, and physiochemical gradients in the complexities of life’s emergence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXosVeiurs%3D&md5=df56923cefb2ca720fc5cbd72c776947CAS |
Stepien P, Johnson GN (2009) Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiology 149, 1154–1165.
| Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXjt1ajs7c%3D&md5=6be1dbf32cadf7f01fe077a9c360d3e6CAS |
Subbarao GV, Ito O, Berry WL, Wheeler RM (2003) Sodium – a functional plant nutrient. Critical Reviews in Plant Sciences 22, 391–416.
Suelter C (1970) Enzymes activated by monovalent cations. Science 168, 789–795.
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Annals of Botany 91, 503–527.
| Na+ tolerance and Na+ transport in higher plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjsVyisbk%3D&md5=88eb811f9dc3f2cd8aa877eaee28c69dCAS |
Trontin C, Tisne S, Bach L, Loudet O (2011) What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Current Opinion in Plant Biology 14, 225–231.
| What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants?Crossref | GoogleScholarGoogle Scholar |
van den Bogaart G, Hermans N, Krasnikov V, Poolman B (2007) Protein mobility and diffusive barriers in Escherichia coli: consequences of osmotic stress. Molecular Microbiology 64, 858–871.
| Protein mobility and diffusive barriers in Escherichia coli: consequences of osmotic stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXlsV2ku7k%3D&md5=a0835a34155054bf33d69ddc7db18dabCAS |
Van Norman JM, Frederick RL, Sieburth LE (2004) BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Current Biology 14, 1739–1746.
| BYPASS1 negatively regulates a root-derived signal that controls plant architecture.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXotFCkur0%3D&md5=8222ebd38728beaa24cc93c030973d24CAS |
Verkman AS (2002) Solute and macromolecule diffusion in cellular aqueous compartments. Trends in Biochemical Sciences 27, 27–33.
| Solute and macromolecule diffusion in cellular aqueous compartments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XktVOmtA%3D%3D&md5=af4af56052bc772a82915fcc3e3d591fCAS |
von Marilaun AK (1896) ‘The natural history of plants.’ (Blackie & Son Ltd: London)
Walch-Liu P, Filleur S, Gan YB, Forde BG (2005) Signaling mechanisms integrating root and shoot responses to changes in the nitrogen supply. Photosynthesis Research 83, 239–250.
| Signaling mechanisms integrating root and shoot responses to changes in the nitrogen supply.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhslGntro%3D&md5=b1eae4292706b3ec884e20cc1ae19a11CAS |
Walia H, Wilson C, Wahid A, Condamine P, Cui X, Close TJ (2006) Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Functional & Integrative Genomics 6, 143–156.
| Expression analysis of barley (Hordeum vulgare L.) during salinity stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhslWgur0%3D&md5=86d62cf263c7fa0a82fa5b1f53dcad18CAS |
Walia H, Wilson C, Ismail AM, Close TJ, Cui XP (2009) Comparing genomic expression patterns across plant species reveals highly diverged transcriptional dynamics in response to salt stress. BMC Genomics 10, 398
Wedel N, Soll J, Paap B (1997) CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proceedings of the National Academy of Sciences of the United States of America 94, 10 479–10 484.
| CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXmt1Gjt7k%3D&md5=5e139a56836fdc20a011018f99336da7CAS |
Welburn JPI, Cheeseman IM (2008) Toward a molecular structure of the eukaryotic kinetochore. Developmental Cell 15, 645–655.
| Toward a molecular structure of the eukaryotic kinetochore.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsVSms7vJ&md5=2d291fc906016ac40b8dec33ede614f5CAS |
Wickens LK, Cheeseman JM (1988) Application of growth analysis to physiological studies involving environmental discontinuities. Physiologia Plantarum 73, 271–277.
| Application of growth analysis to physiological studies involving environmental discontinuities.Crossref | GoogleScholarGoogle Scholar |
Wiggins PM (1990) Role of water in some biological processes. Microbiological Reviews 54, 432–449.
Winkel B (2004) Metabolic channeling in plants. Annual Review of Plant Biology 55, 85–107.
| Metabolic channeling in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXlvFeis70%3D&md5=906016f5c9e8646da8dfd8ee6986a96dCAS |
Wu H-J, Zhang Z, Wang J-Y, Oh D-H, Dassanayake M, Liu B, Huang Q, Sun H-X, Xia R, Wu Y, Wang Y-N, Yang Z, Liu Y, Zhang W, Zhang H, Chu H, Yan C, Fang S, Zhang J, Wang Y, Zhang F, Wang G, Lee SY, Cheeseman JM, Yang B, Li B, Min J, Yang L, Wang J, Chu C, Chen S-Y, Bohnert HJ, Zhu J-K, Wang X-J, Xie Q (2012) Gene complement supports the abiotic stress tolerance phenotype of Thellungiella salsuginea – a genome-based view. Proceedings of the National Academy of Sciences of the United States of America 109, 12219–12224.
| Gene complement supports the abiotic stress tolerance phenotype of Thellungiella salsuginea – a genome-based view.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1GktbvN&md5=d9146f925e4f55a330df8fc14c4b0702CAS |
Xue SW, Yao X, Luo W, Jha D, Tester M, Horie T, Schroeder JI (2011) AtHKT1;1 mediates Nernstian sodium channel transport properties in Arabidopsis root stelar cells. PLoS ONE 6, 9
| AtHKT1;1 mediates Nernstian sodium channel transport properties in Arabidopsis root stelar cells.Crossref | GoogleScholarGoogle Scholar |
Yadav S, Irfan M, Ahmad A, Hayat S (2011) Causes of salinity and plant manifestations to salt stress: a review. Journal of Environmental Biology 32, 667–685.
Yeo A, Yeo M, Caporn S, Lachno D, Flowers TJ (1985) The use of 14C-ethane diol as a quantitative tracer for the transpirational volume flow of water and an investigation of the effects of salinity upon transpiration, net sodium accumulation and endogenous ABA in individual leaves of Oryza sativa L. Journal of Experimental Botany 36, 1099–1109.
| The use of 14C-ethane diol as a quantitative tracer for the transpirational volume flow of water and an investigation of the effects of salinity upon transpiration, net sodium accumulation and endogenous ABA in individual leaves of Oryza sativa L.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2MXlt1Sjtbk%3D&md5=5f33574ce5cfcfe0ddced442623a9c4bCAS |
Yeo AR, Yeo ME, Flowers TJ (1987) The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. Journal of Experimental Botany 38, 1141–1153.
| The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1cXlt1ylug%3D%3D&md5=4301dbf3525f27526ad6939f377203c1CAS |
Yeo AR, Yeo ME, Flowers SA, Flowers TJ (1990) Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theoretical and Applied Genetics 79, 377–384.
| Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance.Crossref | GoogleScholarGoogle Scholar |
Zhang JL, Flowers TJ, Wang SM (2010) Mechanisms of sodium uptake by roots of higher plants. Plant and Soil 326, 45–60.
| Mechanisms of sodium uptake by roots of higher plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsFGrt7vO&md5=17ad0149e5f44ab79b485491eefa7ba1CAS |
Zhu JK (2001) Plant salt tolerance. Trends in Plant Science 6, 66–71.
| Plant salt tolerance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXlsFyjtLs%3D&md5=36de2a21d4f2c605397e01ce5240d467CAS |


