Genetic suppression of plant development and chloroplast biogenesis via the Snowy Cotyledon 3 and Phytochrome B pathways
Diep Ganguly A , Peter Crisp A , Klaus Harter B , Barry J. Pogson A and Verónica Albrecht-Borth A CA ARC (Australian Research Council) Centre of Excellence in Plant Energy Biology, Research School of Biology, Australian National University Canberra, Acton, ACT 0200, Australia.
B Zentrum für Molekularbiologie der Pflanzen, Plant Physiology, University of Tübingen, 72076 Tübingen, Germany.
C Corresponding author. Email: veronica.albrecht@anu.edu.au
Functional Plant Biology 42(7) 676-686 https://doi.org/10.1071/FP15026
Submitted: 2 February 2015 Accepted: 2 April 2015 Published: 18 May 2015
Abstract
Plant development is regulated by external and internal factors such as light and chloroplast development. A revertant of the Arabidopsis thaliana (L.) Heyhn. chloroplast biogenesis mutant snowy cotyledon 3 (sco3–1) was isolated partially recovering the impaired chloroplast phenotype. The mutation was identified in the Phytochrome B (PhyB) gene and is a result of an amino acid change within the PAS repeat domain required for light-induced nuclear localisation. An independent phyB-9 mutation was crossed into sco3–1 mutants, resulting in the same partial reversion of sco3–1. Further analysis demonstrated that SCO3 and PhyB influence the greening process of seedlings and rosette leaves, embryogenesis, rosette formation and flowering. Interestingly, the functions of these proteins are interwoven in various ways, suggesting a complex genetic interaction. Whole-transcriptome profiling of sco3–1phyB-9 indicated that a completely distinct set of genes was differentially regulated in the double mutant compared with the single sco3–1 or phyB-9 mutants. Thus, we hypothesise that PhyB and SCO3 genetically suppress each other in plant and chloroplast development.
Additional keywords: Arabidopsis thaliana, chloroplast development, gene regulation phytochrome interacting factor 4.
Introduction
Chloroplast development in plants is influenced by a variety of environmental cues such as light and abiotic stress. Although light is required to initiate chloroplast biogenesis (Pogson and Albrecht 2011), abiotic stress has been demonstrated to have a negative impact on chloroplast biogenesis and function (Meskauskiene et al. 2001; Dutta et al. 2009; Kim et al. 2009; Dalal and Tripathy 2012). Overall, plant development and chloroplast development are tightly connected as each process is affected by impairment in the other. For example, impaired or even just delayed chloroplast development affects flowering time, biomass production and seed set.
One primary environmental factor influencing seedling development is the perception of light by different photoreceptors such as the two major protein families, phytochromes and cryptochromes (Sullivan and Deng 2003). Whereas cryptochromes are involved in the perception of blue light, phytochromes are involved in red and far-red light-mediated signalling (Reed et al. 1993; Sullivan and Deng 2003). In Arabidopsis thaliana (L.) Heynh., five different phytochromes have been identified, with Phytochrome A and Phytochrome B (PhyB) being the most prominent in light-mediated signalling, as well as two cryptochromes, Cryptochrome 1 and Cryptochrome 2.
Phytochromes, upon perception of light, undergo structural changes from the inactive Phy in red light (Pr) form to the active Phy in far-red light (Pfr) form, which is subsequently transferred into the nucleus (Chen et al. 2005). Here, they are involved in regulating the activity of transcription factors (Castillon et al. 2007; Waters et al. 2009). In particular, one group of transcription factors was found to play an important role during seedling establishment and photomorphogenesis in regulating important genes that encode proteins for gibberellic acid biosynthesis and signalling, or chlorophyll biosynthesis (Castillon et al. 2007). This class of transcription factors has been termed phytochrome interacting factors (PIFs), for which at least five members have been described in A. thaliana (Castillon et al. 2007). Interestingly, interaction of PhyB with the different PIF proteins mostly results in their phosphorylation, which then targets the PIF proteins for ubiquitination-mediated protein degradation (Shin et al. 2009). Additionally, the developmental stage of the plant effects the functional interaction between PhyB and PIF (Shin et al. 2009; Stephenson et al. 2009). The transcriptional networks of the PIF proteins can overlap; however, specific regulatory functions exist: for example, PIF1 has been described as functioning in germination and chlorophyll biosynthesis, and PIF3 in the greening of seedlings (Huq et al. 2004; Monte et al. 2004). Indeed, phyB mutants are pale green throughout the entire plant cycle due to impaired chloroplast development (Chory et al. 1989). In the absence of functional PhyB, the PIF transcription factors are not degraded and thus continue to repress transcription of the essential genes required for chloroplast development and function (Chory et al. 1989; Stephenson et al. 2009). Loss of PhyB function not only impairs chloroplast development but also affects other processes such as shade avoidance, water use efficiency and flowering time (Reed et al. 1993; Boccalandro et al. 2009).
Although chloroplast biogenesis differs between cotyledons and true leaves, some common regulators have been identified in the last decade (Bauer et al. 2001; Pogson and Albrecht 2011). These include proteins that affect gene transcription, protein translation, chloroplast protein import, thylakoid formation or regulation of photosynthesis (Bauer et al. 2001; Sakamoto et al. 2008; Kessler and Schnell 2009; Pogson and Albrecht 2011). We have identified a group of genes whose impact on chloroplast biogenesis is greater in cotyledons than leaves. These genes have been named Snowy Cotyledon (SCO) (Albrecht et al. 2006; Albrecht et al. 2008; Pogson and Albrecht 2011; Tanz et al. 2012; Albrecht-Borth et al. 2013). The snowy cotyledon 3 (sco3) mutant was of particular interest, encoding a protein of unknown function that is not located in the chloroplasts but in the periphery of peroxisomes. Dysfunction of this protein impairs the fine structure of the cytoskeleton (Albrecht et al. 2010). In contrast, the other identified SCO proteins are required for chloroplast protein translation, as is the case for SCO1; chloroplast protein folding and targeting, as shown for the protein disulfide isomerase SCO2, or a likely novel protease activity, as is the case for SCO4 (Albrecht-Borth et al. 2013; Pogson and Albrecht 2011; Tanz et al. 2012). Although the effect of the sco3 mutation on chloroplast development in cotyledons is apparent, only a few genes have been found to be misregulated in the sco3 mutant (Albrecht et al. 2010).
Here we describe the identification of a revertant of sco3–1 that is due to a mutation in the PhyB gene. We propose a mechanism of crosstalk possibly occurring between the phytochrome-mediated signalling pathway and SCO3-mediated processes, and investigate how they alter nuclear gene transcription to complement impaired chloroplast biogenesis.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh plants were grown either on sterile MS medium (Murashige and Skoog 1962) with 0.8% agar or on soil under normal long-day conditions (16 h light, 8 h dark) at 21°C. For sterile growth conditions, seeds were surface sterilised using 70% ethanol and washed three times with autoclaved bidistilled water before plating. After overnight stratification, the seeds were transferred to a room with a constant temperature and the described growth conditions.
Analyses of TILLING lines
The SCO3 genomic sequence was submitted to the A. thaliana TILLING project (http://tilling.fhcrc.org/files/Welcome_to_ATP.html) to identify additional mutant alleles in SCO3. Seeds that should contain mutations according to the TILLING results were obtained from Arabidopsis Biological Research Centre (ABRC), sown on soil and monitored for the sco3–1 typical phenotype. For each line, DNA was extracted as described by Albrecht et al. (2006) and the predicted genomic region of the mutation was amplified and subsequently sequenced to verify the mutation. In aim to outcross the erecta mutation in the parental Big Mamma line, the TILLING mutants were crossed several times into A. thaliana cv. Columbia (Col) and rescreened for a pale green phenotype. The segregation of the sco3 mutation with the pale phenotype was analysed by resequencing the gene region.
Isolation of sco3–1 revertants
Approximately 25 000 seeds of sco3–1 were mutagenised with ethyl methyl sulfonate (EMS) as described by Albrecht et al. (2006), and the M2 generation was plated on MS medium and screened for seedlings with an increased green cotyledon phenotype. Mutant lines identified in the M2 generation were rescreened in the M3 generation to verify the observed phenotype. Subsequently, they were backcrossed into sco3–1 to eliminate most of the other mutations caused by ethyl methyl sulfonate and to identify the recessive or dominant nature of the second site mutation. One particular mutant with a long hypocotyl and greener cotyledons than sco3–1 was identified in the obtained revertants. Because of its particularly phyB-like phenotype, the PhyB gene was amplified and sequenced to identify potential mutations in this gene. Since it turned out that the mutation in this mutant is dominant, we did not proceed to isolate the single phyB-XD mutation but introduced a recessive allele of phyB into sco3–1. The second mutant allele of phyB, phyB-9, was crossed into sco3–1 to confirm the observed phenotype and mutation of the identified revertant.
Chlorophyll analysis
For chlorophyll quantification 30 mg of 7-day-old seedlings grown on MS medium under 16 h light conditions were harvested, frozen and ground in liquid nitrogen and resuspended in 80% acetone. The measurements were performed on a plate reader (Infinite M1000 PRO, TECAN, Switzerland) for optical density (OD)663 and OD645, and the calculations were executed as described by Albrecht et al. (2006).
Scanalyzer analysis
The analysis of the greenness in rosette leaves and the circumference of the rosettes in 3-week-old plants was performed using the Scanalyzer system (LemnaTec, Germany). For this analysis, plants were grown on trays of soil for 3 weeks (or longer for the analysis of flowering time) and were transferred under the camera of the Scanalyzer image capture system. Subsequent image analysis was performed according to the Scanalyzer manual (LemnaTec).
Microarray analyses and quantitative real-time PCR
Analysis of the differences in the gene expression profiles between Col, sco3–1, phyB-9 and sco3–1phyB-9 was performed using the Gene Chip Arabidopsis ATH1 genome arrays (Affymetrix, CA, USA) at the Ramaciotti Centre (Sydney, NSW, Australia) in biological triplicate. Total mRNA of 4-day-old seedlings grown under long day conditions (16 h light : 8 h dark) was isolated and the DNA was treated using the Spectrum Total RNA Kit (Sigma, St Louis, MO, USA). Quality check metrics were provided by the Ramaciotti Centre. The CEL files obtained were further analysed, including additional quality check analyses using the Partek Genomic Suite (http://www.partek.com/pgs, accessed 17 April 2015). Log-transformed (log base 2) data were analysed to detect differentially expressed genes between each line using a one-way ANOVA based on the method of moments (Eisenhart 1947) and Fisher’s LSD contrast method for differential regulation between any two of the lines using the work-flow of the Partek Genomic Suite. Significantly misregulated genes had to be misregulated by at least twofold (up or down) using a Benjamini step-up adjusted P-value with false discovery rate of 0.05. Gene name and function were obtained from the The Arabidopsis Information Resource (TAIR) database (www.arabidopsis.org). Of particular interest were the genes that were specifically up- or downregulated in the sco3–1phyB-9 double mutant. The microarray data produced and discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible via the Gene Expression Omnibus series accession number GSE67168 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67168).
For verification of the observed changes in gene expression in the microarray analysis, select genes of interest were analysed using quantitative real-time PCR. These genes were Ferritin 1 (FER1, At5g01600), Early Light-Induced Protein 1 (ELIP1, At3g22840), Flowering Locus C (FLC, At5g10140), PIF4 (At2g43010) and HEME2 (encoding uroporphyrinogen decarboxylase, At2g40490). For this, total mRNA of each of the four lines used in the microarray study was reverse transcribed using the Superscript III cDNA Synthesis Kit (Invitrogen, The Netherlands). Relative transcript abundance was quantified in Col, sco3–1, phyB-9 and sco3–1phyB-9 plants, using the SYBR Green JumpStart Taq Ready-Mix (Sigma-Aldrich, MO, USA). Three technical replicates, one for each of three biological replicates for each line, were performed using the Light Cycler 480 (Roche, Switzerland). The obtained data were analysed by relative quantification (quantitative cycle (Cq) values) using the Protein Phosphatase 2A gene (At1g13320) as a housekeeper control.
Gene Ontology annotation analysis was performed using the TAIR database and the subsequent data were analysed according to Narsai et al. (2007).
Generation of overexpression lines
The cDNA of PIF4 (At2g43010) was cloned into the binary vector pMDC85 under the control of the 35S promoter, transformed into Col and sco3–1 plants, and selected on hygromycin. Since we did not obtain any transgenic plants for the PIF4 overexpression construct in sco3–1, several independent transformed lines of Col containing the PIF4 overexpression construct were crossed with sco3–1. The subsequent F2 generation was screened for plants harbouring both the PIF4 overexpression construct as well as being homozygous for sco3–1.
Results
Alellic SCO3 point mutations demonstrate the involvement of SCO3 in chloroplast development at all stages of plant development
Null alleles of SCO3 are lethal in the embryo and the sco3–1 allele results in pale green cotyledons (Albrecht et al. 2010). Newly identified sco3–5 and sco3–6 alleles recovered from a TILLING screen resulted in both pale green cotyledons and true leaves (Fig. S1, available as Supplementary Material to this paper). Thus, SCO3 is essential for plastid development at all stages of a plant’s life cycle, with the mutation’s penetrance determining the degree of deleterious impacts at different developmental stages.
Mutations in the PhyB gene rescue specific sco3–1 phenotypes and vice versa
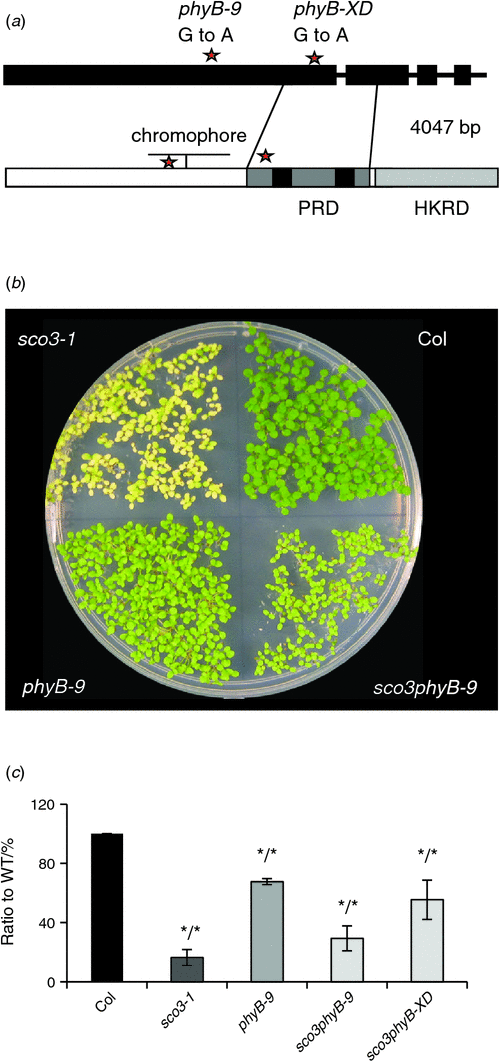
In order to determine how the peroxisomal-associated SCO3 protein impacts chloroplast biogenesis, a second-site mutagenesis was performed on 25 000 sco3–1 seeds. The M2 population was screened for reversions of the sco3–1 bleached cotyledon phenotype (i.e. plants that exhibited green cotyledons). Several of these mutants were identified and one of these had a long hypocotyl. Since it is known that phyB mutants exhibit a long-hypocotyl phenotype, the coding region of PhyB in this revertant was amplified, sequenced and shown to have a G to A mutation, which resulted in an amino acid change from alanine to threonine at Position 663 (A663T) (Fig. 1a and Fig. S2b). This amino acid is conserved in all phytochromes and is located at the beginning of the PAS repeat domain, which is required for nuclear localisation of the PhyB protein (Chen et al. 2005; Krall and Reed 2000). Since we observed a segregation of green and pale seedlings in the M2 generation, it is to be expected that the phyB mutation in this line is dominant. Subsequently, this revertant was termed sco3–1phyB-XD.
|
To determine the nature of the phyB-XD mutation, the sco3–1phyB-XD double mutant was crossed with sco3–1. In the F2 population, 813 seedlings were counted, where 615 (76%) were green and 198 (24%) were pale. This 3 : 1 segregation indicated that the phyB-XD mutation does indeed revert the sco3–1 phenotype in a dominant manner. To further confirm the role of PhyB in the reversion, sco3–1 was crossed with the recessive phyB-9 allele with a premature stop codon (Fernandez et al. 2005) and the homozygous double mutant line was isolated in the F2 generation. Introducing the recessive phyB-9 allele into the sco3–1 mutant background also resulted in a partial greening of the pale cotyledons, as was observed in sco3–1phyB-XD (Fig. 1b). Comparison of the chlorophyll content of the double mutant with sco3–1 and Col showed that sco3–1phyB-9 has about twice as much chlorophyll as sco3–1 in 7-day-old seedlings, whereas the initially identified phyB-XD allele almost tripled the amount of chlorophyll compared with sco3–1 (Fig. 1c, Table 1). Thus mutations in the PhyB gene can partially rescue the pale green sco3–1 phenotype.
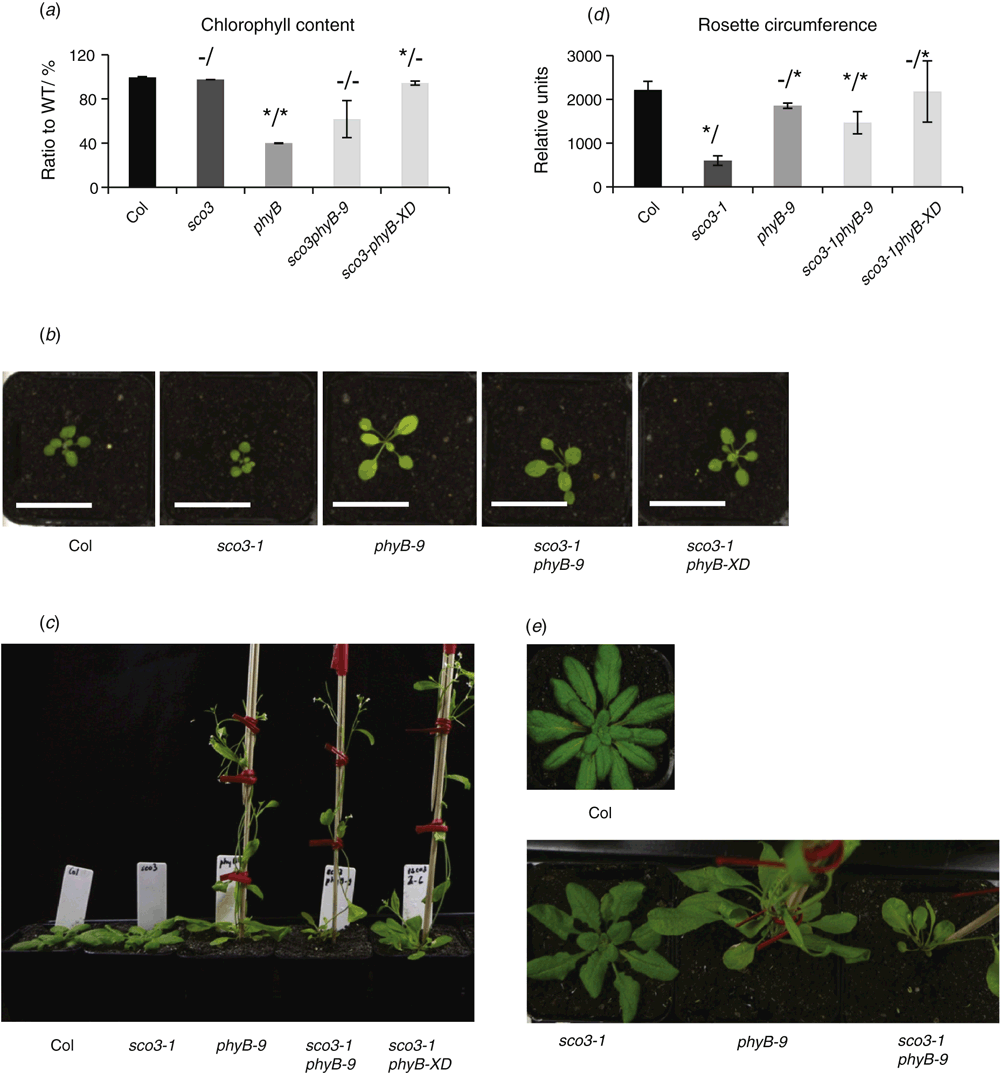
Similarly, although phyB-9 mutant lines exhibited a pale true-leaf phenotype, quantification of the chlorophyll content of the leaves in 3-week-old plants showed that the introduction of the sco3–1 mutation partially recovered the chloroplast development in true leaves (Fig. 2a, b, Table 1).
With respect to flowering time, combining the phyB-9 and sco3–1 mutations tended to enhance the early flowering phenotype normally observed in phyB-9. In other words, both the phyB-9 mutant (9.5 ± 1.9 rosette leaves, 4 weeks) and sco3–1phyB-9 double mutant (5 ± 0.8 rosette leaves, 3.5 weeks) started flowering earlier under long-day conditions compared with Col (12.3 ± 0.9 rosette leaves; 6 weeks) and sco3–1 (>16 rosette leaves, 8–10 weeks) (Fig. 2c, Table 1). The rosette circumference of 3-week-old sco3–1phyB-9 plants is smaller than that of phyB-9 but bigger than that of the sco3–1 mutant plants, whereas sco3–1phyB-XD resembles wild-type plants in circumference and has wild-type green leaves with longer petioles (Fig. 2d, Table 1). With respect to leaf morphology, the introduction of the sco3–1 mutation into phyB-9 also led to enhanced defects: whereas both sco3–1 and phyB-9 were able to form large rosettes the double mutant had very few rosette leaves (Fig. 2e, Table 1).
Sco3–1 does not alter the hypocotyl length of seedlings grown under red light
An important function for PhyB is light-mediated photomorphogenesis under red light. Mutants impaired in phyB not only exhibit a longer hypocotyl under white light but are also insensitive to red light (Mira-Rodado et al. 2007). Therefore, an analysis was performed by germinating Col, sco3–1, phyB-9 and the double mutant sco3–1phyB-9 under increasing red light conditions. Whereas Col and sco3–1 demonstrated a decrease in hypocotyl length with increased red light intensities, neither phyB-9 nor sco3–1phyB-9 responded to red light (Fig. S3). Thus the introduction of the sco3–1 mutation into phyB-9 does not influence the hypocotyl growth of the seedlings under red light.
SCO3 influences post-germination chloroplast development via an embryogenesis-linked mechanism
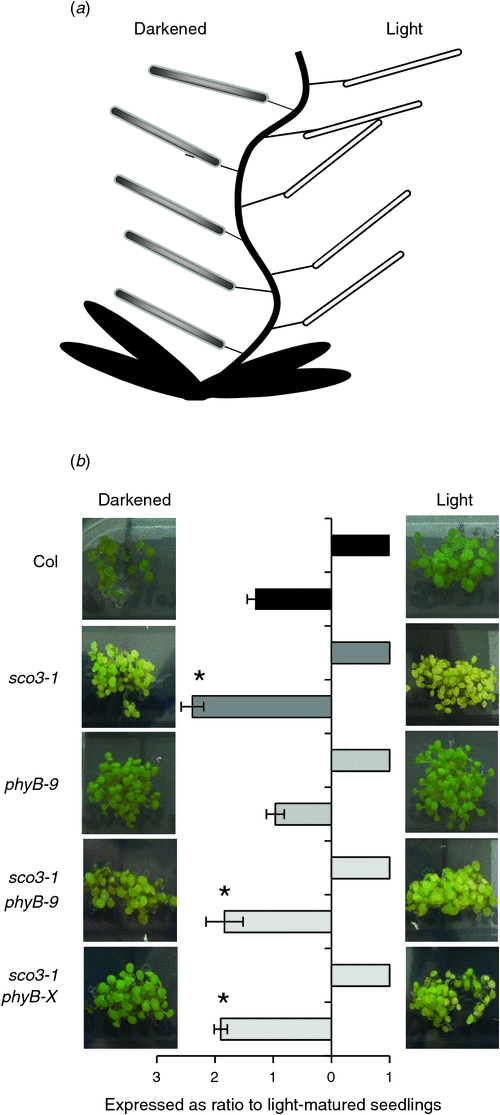
In a recent publication, it was shown that light during embryogenesis positively affects chloroplast development after germination in the ex1ex2 double mutant (Lee et al. 2007). Since both of the single mutants, sco3–1 and phyB-9, are impaired in chloroplast development in young seedlings, and PhyB is involved in light perception, a light-induced defect in chloroplast formation during embryogenesis cannot be excluded. To test this, every second silique of the main bolting stem was covered with aluminium foil for dark embryo maturation, whereas the other siliques were used as controls for embryo development in the light (Fig. 3a). Mature seeds were harvested at the same time, surface sterilised and sown on MS medium, and the chlorophyll content of 7-day-old seedlings was determined. Although there was no significant change in the phyB-9 mutant, chlorophyll content was unexpectedly enhanced threefold in the sco3–1 mutant and twofold in both double mutants, which suggests that the effect of the sco3–1 mutation is mitigated by the absence of light during embryogenesis (Fig. 3b, Table 1).
|
A distinct set of genes is differentially regulated in sco3–1phyB-9 compared with either sco3–1 or phyB-9
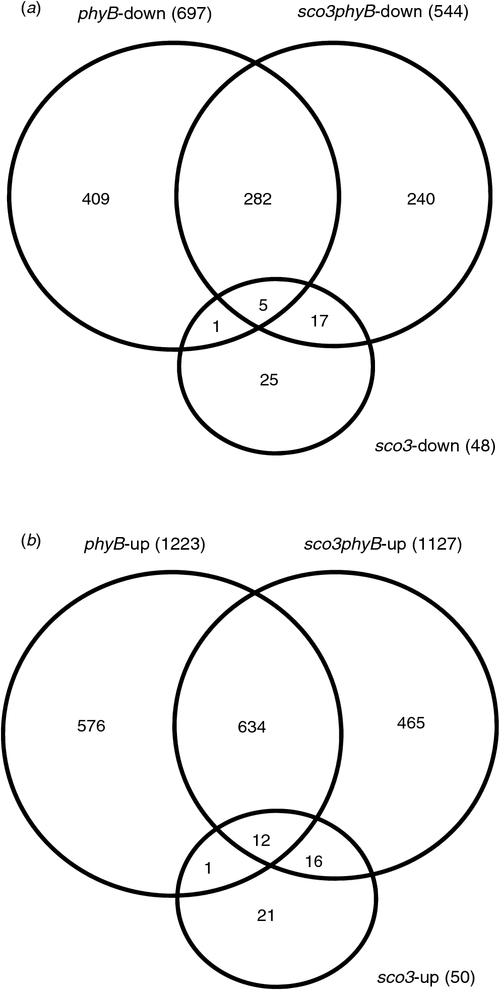
In order to investigate the opposing impacts of SCO3 and PhyB mutations in the greening of young seedlings, we measured the global transcript levels of nuclear-encoded genes in 4-day-old seedlings grown under long-day conditions. It is known that PhyB regulates the transcription of many nuclear-encoded genes (Sullivan and Deng 2003; Monte et al. 2004; Moon et al. 2008; Shin et al. 2009), whereas only a small number of transcripts are altered in young sco3–1 seedlings grown under continuous light (Albrecht et al. 2010). We performed a new transcriptome analysis on 4-day-old seedlings using Affymetrix chips and compared the transcript levels of Col with those of the single and double mutants. As expected, many genes were downregulated (697) or upregulated (1223) in the phyB-9 single mutant and only 50 genes were upregulated and 48 genes were downregulated in sco3–1 (Fig. 4a and Table S3).
|
Interestingly, in the sco3–1phyB-9 double mutant, 544 transcripts were downregulated and 1127 were upregulated. Volcano plots were made to compare the distribution of misregulated transcripts between the different genotypes (Fig. S4). From these plots, it is evident that many genes were uniquely misregulated in sco3–1phyB-9 (green dots in Fig. S4a, b and Fig. 4a). Although some of the genes were specific for sco3–1 (blue dots in Fig. S4a), introduction of the phyB-9 mutation reverted the expression of 25 out of 48 downregulated genes and 21 out of 50 upregulated genes in sco3–1.There was also a considerable proportion of genes misexpressed in phyB-9 (blue dots in Fig. S4b) that were not so in the double mutant (Fig. 4a, green dots in Fig. S4b). In addition, half of the genes misexpressed in the double mutant were unique to the double mutant compared with phyB-9 and sco3–1 (Fig. 4a, b, Table S2; genes misregulated in sco3–1 are listed in Table S3). A question is whether the nonoverlapping genes between single and double mutants correspond to genes that narrowly miss significance criteria. The volcano plots do not support this hypothesis, as the distribution of 900 unique PhyB (blue dots) largely overlies the >20 000 unchanged transcripts (black dots) in the double mutant (Fig. S4b).
Analysis of the ontological (Gene Ontology) annotation of the genes that were uniquely up- or downregulated in the sco3–1phyB-9 double mutant compared with the single mutants indicated an over-representation of plastid or nucleus-localised proteins compared with the whole genome distribution. Proteins of the cell wall and the extracellular compartment were over-represented in the downregulated genes in the double mutant. In the upregulated genes, an over-representation of proteins involved in abiotic and biotic stress responses were observed, whereas in both the up- and downregulated genes, protein metabolism was under-represented (Table S4). The latter seems to balance out the overproduction of proteins in sco3–1, which demonstrates the impairment in chloroplast development and thus results in greener seedlings. A similar observation has been made in revertants of the chloroplast development mutants variegated, where a reduction in chloroplast protein biosynthesis counterbalanced the deficiency in chloroplast protein degradation, resulting in the greening of the plants (Miura et al. 2007). However, the most pronounced is the expression of genes in both gene pools that are involved in abiotic and biotic stress responses, indicating that continuous stress was present in the double mutant.
As sco3-1phyB-9 is a mutant with a partially recovered chloroplast development phenotype, we were interested to discover whether within the pool of genes uniquely up- or downregulated, there were genes that are known to be involved in or to affect chloroplast development. Indeed, when we looked at the list of genes that were upregulated in the double mutant and predicted to be localised in the chloroplast, many of the genes with known function were involved in the regulation of transcription or post-transcriptional processing, or protein translation such as the pentatricopeptide repeat (PPR) protein Early Chloroplast Biogenesis 2 or the RNA-processing protein High Chloroplast Fluorescence 152 (Meierhoff et al. 2003; Cao et al. 2011). Genes involved in chloroplast protein import are upregulated such as Translocon at the outer envelope membrane of chloroplasts 55 (TOC55) or Chloroplast import apparatus 2 (CIA2) and CIA2-like, both of which upregulate the transcription of the chloroplast import apparatus proteins TOC33 and TOC75 (Sun et al. 2009). Several genes were identified in the defective embryo screen, suggesting that an upregulation of these genes might benefit chloroplast development. In contrast, in the list of downregulated genes, only a few are with proteins of known functions such as the chlorophyll biosynthesis protein HEME2.
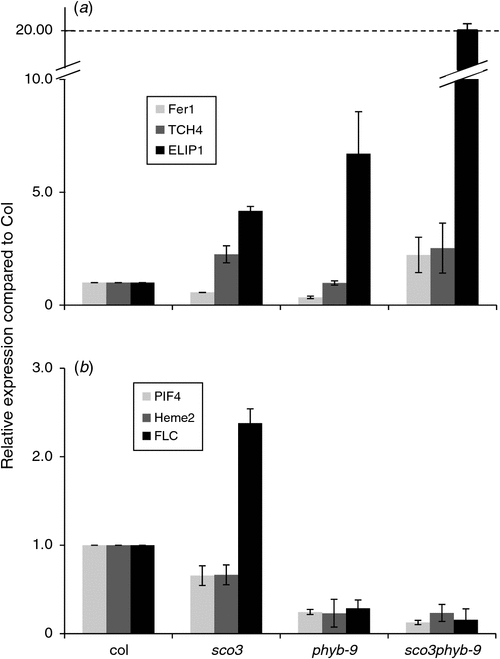
Interestingly, upon closer examination of the misregulated genes and their encoded protein functions, it was observed that several gene families are over-represented in the list to a greater degree than expected, such as the upregulation of UDP glycosyltransferases involved in secondary metabolism (Ross et al. 2001), or the downregulation of proteins involved in cell wall metabolism such as the ketoacyl CoA synthases involved in long chain fatty acid elongation (Haslam and Kunst 2013), xyloglucan transferases and hydrolases (Rose et al. 2002), arabinogalactan proteins or extensins (Table S2). To confirm the results obtained from the microarray data, quantitative real-time PCR on RNA extracted from the different lines was performed using cDNA-specific primers for selected genes that were either up- or downregulated in sco3–1phyB-9 (Fig. 5a, b, Table S5). In summary, FER1, Touch 4 (TCH4) and ELIP1were all upregulated, and PIF4, HEME2 and FLC were all downregulated in sco3–1phyB-9. Except for TCH4, which was downregulated in the array, the data from the quantitative PCR confirmed (FER1, FLC) the array data or showed an enhanced up- or downregulation (ELIP1, PIF4 and HEME2).
|
Overexpression of PIF4 is able to partially rescue the sco3–1 pale cotyledon phenotype
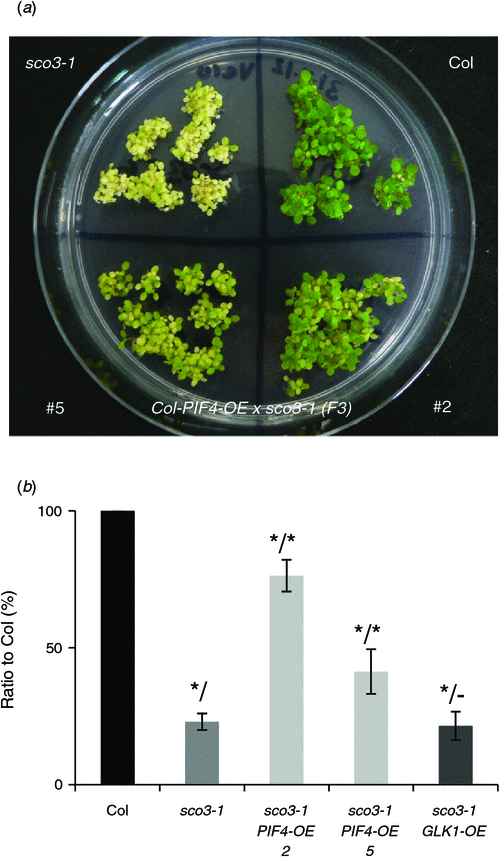
Our previous transcript analysis on sco3–1 revealed that very few transcription factors are downregulated, such as Golden-like 1, which encodes a protein involved in the regulation of many photosynthesis genes, and PIF4 (Albrecht et al. 2010). PIF proteins interact with, are regulated by and regulate PhyB. Under continuous light, the transcript levels of PIF4 in sco3–1 seedlings was downregulated five fold and, in this analysis under long-day conditions, by –1.5 fold (Fig. 5) (Albrecht et al. 2010). Since PIF proteins are regulated by phytochrome proteins in a light-dependent manner (Castillon et al. 2007), we reasoned that overexpression of PIF4 might complement sco3–1. Seedlings transformed with the PIF4 overexpression construct resulted a similar greening of the cotyledons to that observed for the sco3–1phyB double mutants (Fig. 6a, b). Quantification of PIF4 expression in these lines demonstrated an upregulation of almost twofold in transgenic lines compared with Col and fourfold compared with sco3–1 (PIF4 overexpression in sco3–1 #2: 1.86 ± 0.68; PIF4 overextression in sco3–1 #5: 1.75 ± 0.9). Thus the PIF4 protein seems to affect the regulation of chloroplast development and overexpression improves chloroplast biogenesis in sco3–1. However, when one quantifies PIF4 transcript levels in sco3–1phyb-9, in both the microarrays and quantitative PCR there is, in fact, a downregulation rather than the expected upregulation of PIF4. This result somewhat contradicts our observation of sion of pale cotyledon phenotype in sco3–1 by overexpression of PIF4. This suggests a complex regulation that will require further investigation.
|
Discussion
In this study, we observed that the introduction of mutations into the PhyB gene in the sco3–1 mutant background resulted in a partial recovery of the pale cotyledon phenotype in sco3–1 but also demonstrated complex traits in phenotypes that can only be explained by the differential additive or suppressive genetic regulation summarised in Table 1, which will be discussed in more detail. These genetic networks reveal a complex interaction between the SCO3 and PHYB pathways to regulate plant development and chloroplast biogenesis.
SCO3 and PhyB regulate a novel set of genes during seedling chloroplast biogenesis
Loss of normal SCO3 protein function results in impaired development of chloroplasts from their progenitors, the proplastids, by affecting the fine structure of the cellular actin cytoskeleton and impairing the adaptation of the plants to extreme CO2 concentrations (Albrecht et al. 2010). The sco3–1 mutant was reported to have no observable phenotype in the rosette leaves under normal growth conditions, whereas other newly identified mutations, sco3–5 and sco3–6, resulted in a pale green adult leaf phenotype, indicating a role for SCO3 throughout plant development in photosynthetic tissues.
The loss of PhyB function in the sco3–1 mutant resulted in a greening of the cotyledons, indicating improved chloroplast biogenesis (Fig. 1b). This might indicate that SCO3 genetically suppresses PhyB. On the other hand, the combination of both mutations results in an intermediate phenotype, since both single mutants have deleterious mutations that result in pale embryonic leaves. Analysis of the transcriptome of the double mutant revealed that, in addition to an overlap of ~50% with up- or downregulated genes identified in either phyB-9 or sco3–1, a completely distinct set of genes was differentially expressed in the double mutant (Fig. 4). The phyB-9 mutant is a signalling mutant that impairs the transcription of many genes, such as photosynthesis genes, by interaction with the PIF transcription factors. This is reflected in our transcriptomic data, where almost 2000 transcripts were found to be differentially expressed in this mutant. On the other hand, the transcriptome analysis in sco3–1 indicates that only a few genes were differentially expressed in the mutant compared with the wild-type, despite displaying impaired chloroplast biogenesis. SCO3 can only indirectly influence the transcription of genes, as it is located in the peroxisome border. With the data from sco3–1, we can assume that even though chloroplast development is severely impaired in the single mutant, the majority of genes required for chloroplast development are being expressed but some other function for chloroplast development is impaired in the sco3–1 mutant. The observations of the transcript regulation in the sco3–1phyB-9 double mutant indicate that the observed changes of this subset of genes, which are differentially expressed in the double but not in the single mutants, result in an improvement in chloroplast development. Furthermore, the overexpression of one PhyB interacting factor, PIF4, in sco3–1 resulted in a similar greening of the cotyledons to that observed in the sco3–1phyB-9 mutant. This might indicate that PIF4, which regulates and is regulated by the function of PhyB (Leivar et al. 2008; Shin et al. 2009), is involved in integrating the regulatory pathways of SCO3 and PhyB. So far, no direct involvement in regulating chloroplast biogenesis has been demonstrated for PIF4, though its function in integrating diverse environmental signals has been discussed (Lucyshyn and Wigge 2009).
We investigated a set of genes that were differentially regulated in the double mutant and verified their expression in the single and double mutants. These genes are either involved in development, in cellular organisation and biogenesis (TCH4, HEME2 and FLC) or in stress responses (FER1, ELIP1). FER1 was demonstrated to be regulated by hydrogen peroxide in several stress analyses (op den Camp et al. 2003; Rossel et al. 2007; Gordon et al. 2012). In this study, FER1 was observed to be downregulated in the single mutants; however, it was upregulated in the sco3–1phyB-9 double mutant (Fig. 6a), suggestive of increased levels of or sensitivity to reactive oxygen species. ELIP1 demonstrated an additive effect in gene regulation, as its transcript levels were four- to fivefold higher in the double mutant as they were in either of the single mutants, sco3–1 and phyB-9. ELIP1 is located at the thylakoids and is involved in the greening of etiolated seedlings under continuous high light (Casazza et al. 2005). Indeed, a photoprotective function of ELIP1 has been identified as accumulating linearly with light intensity and correlates to the degree of photoprotection in PSII (Heddad et al. 2006), which could suggest a role for ELIP1 in partial complementation.
Loss of SCO3 function dominates in the regulation of embryonic factors required for seedling chloroplast development
The covering of siliques resulted in the recovery of the delayed chloroplast biogenesis in the ex1ex2 double mutant, which is impaired in its ability to relay singlet oxygen-mediated signalling from the chloroplast to the nucleus (Kim et al. 2009). Darkening of the embryos interrupts some chloroplast functions such as photosynthesis and might affect not only gene transcription but also the assimilation of the different compounds normally produced in embryos exposed to light. Since PhyB is a major factor required for the perception of light and the subsequent regulation of many photosynthesis genes, we performed a similar experiment with our mutant lines to analyse the effect of darkness during embryogenesis on the greening process of the emerging seedlings. Darkening the embryos during maturation resulted in seedlings having increased chlorophyll levels except for the phyB-9 mutant, indicating the importance of PhyB for light perception in the embryos (Fig. 3b, Table 1).
The greening effect of dark embryo maturation is enhanced in the lines containing the sco3–1 mutation giving further evidence that SCO3 protein function is required for chloroplast development. What is curious is that the developing embryos of sco3–1 are as green as those in the wild-type (Albrecht et al. 2010) but if they mature in the dark, the resulting seedlings are greener than if they mature in the light. This suggests an unknown embryonic factor will be required for chloroplast development in germinating seedlings. In their analysis on their double mutant ex1ex2, Kim et al. (2009) revealed that the influence of photosynthesis-derived singlet oxygen generated during embryogenesis has an impact on chloroplast development after germination. Since no singlet oxygen-specific genes are differentially regulated in the sco3–1 mutant (as discussed in Albrecht et al. 2010) and there did not seem to be an overlap of genes differentially regulated between ex1ex2 and sco3–1phyB-9, we assume that in sco3–1 or sco3–1phyB-9, this type of signalling is not involved in the regulation of chloroplast development. Our analysis indicates that PhyB is involved in the formation of this yet unknown embryonic factor(s) as it is required for the increase of chlorophyll and thus improved chloroplast formation in the emerging seedlings.
Loss of PhyB function dominates in the regulation of flowering time
In addition to chloroplast development, SCO3 and PhyB also influence other aspects of plant development. PhyB regulates flowering time, since it is part of the signalling system entraining the circadian clock (Sullivan and Deng 2003). Loss of the PhyB signalling pathway results in early flowering plants, which is even further enhanced by introducing the sco3–1 mutation (Table 1). The mutation in sco3–1 appears to result in delayed flowering and upregulation of FLC. That is, FLC is upregulated in sco3–1 but downregulated in phyB-9 and is even further upregulated in sco3–1phyB-9, which correlates with the observed flowering phenotype for each single and double mutant (Figs 2a and 6b). Indeed, a correlation between an abundance of mRNA and flowering time has been demonstrated by other groups, where a reduced amount of mRNA of the MADS-box transcription factor encoding gene FLC resulted in an earlier flowering of the plants (Swiezewski et al. 2007).
Interestingly, the formation of the typical morphology of a rosette in A. thaliana is dramatically impaired in the sco3–1phyB-9 double mutant (Fig. 2, Table 1), which appears to be an additive genetic interaction, resulting in a phenotype that is more severe than the two single mutants. In the phyB mutant, a reduction of rosette leaves was observed, which was thought to be linked to the early flowering phenotype. This was only observed in the sco3–1phyB-9 mutant and not in the sco3–1phyB-XD double mutant. Thus the inference is that this phenotype is influenced by the mutant allele, which, in phyB-9, is close to the binding region of the phytochromobilin chromophore; in phyB-XD, the mutation is within the PAS repeat domain close to the first PAS motif (Krall and Reed 2000). The latter was found to be required for nuclear relocalisation of the PhyB protein whereas phytochromobilin itself is necessary for the perception of light.
The analyses of the sco3–1, phyB-9, and sco3–1phyB lines revealed that a complex additive and suppresive genetic regulation of the two SCO3- and PhyB-dependent networks interferes in regulating different plant developmental stages (Table 1). The global expression data are consistent with this complexity and reveal similar proportions of coexpressed and uniquely expressed genes in sco3–1phyB-9 compared with phyB-9. These analyses, while revealing novel aspects of the regulation of plant development, raise new questions. For example, what are the other factors in the integration of developmental process? What factors are generated during embryo development that influence chloroplast development in seedlings? Why is there a differential regulation between all the processes analysed during plant development?
SCO3 is an essential gene for embryogenesis and chloroplast development with an as yet unidentified function. We have shed new light on the role of SCO3 by identifying its novel but complex genetic interaction with PhyB and the canonical light-mediated transcriptional regulation pathway in plant development encompassing not only chloroplast development and the involvement of embryonic factors but also flowering time and rosette formation.
Acknowledgements
We thank Estee Tee and Lee Constable for their assistance in growing and harvesting the plants. This project has received support from the Australian Research Council Centre of Excellence in Plant Energy Biology (CE0561495).
References
Albrecht V, Ingenfeld A, Apel K (2006) Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Molecular Biology 60, 507–518.| Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XitFOhs7k%3D&md5=67026d996160b386eab9d9b404076eaeCAS | 16525888PubMed |
Albrecht V, Ingenfeld A, Apel K (2008) Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Molecular Biology 66, 599–608.
| Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXjsFWlsrw%3D&md5=5016368e71d2be803acf5b9a3e584265CAS | 18209955PubMed |
Albrecht V, Simkova K, Carrie C, Delannoy E, Giraud E, Whelan J, Small ID, Apel K, Badger MR, Pogson BJ (2010) The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. The Plant Cell 22, 3423–3438.
| The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsF2ms77M&md5=d47c31b22e950c1965e2d4ba34fc27ffCAS | 20978221PubMed |
Albrecht-Borth V, Kauss D, Fan D, Hu Y, Collinge D, Marri S, Liebers M, Apel K, Pfannschmidt T, Chow WS, Pogson BJ (2013) A novel proteinase, SNOWY COTYLEDON4, is required for photosynthetic acclimation to higher light intensities in Arabidopsis. Plant Physiology 163, 732–745.
| A novel proteinase, SNOWY COTYLEDON4, is required for photosynthetic acclimation to higher light intensities in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhs1OjsLzM&md5=33daf2ad85129a0e6f0f2ce37770343cCAS | 23940253PubMed |
Bauer J, Hiltbrunner A, Kessler F (2001) Molecular biology of chloroplast biogenesis: gene expression, protein import and intraorganellar sorting. Cellular and Molecular Life Sciences 58, 420–433.
| Molecular biology of chloroplast biogenesis: gene expression, protein import and intraorganellar sorting.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXjtVahtLw%3D&md5=b13d33f1dde072af2733c94fa855d29dCAS | 11315189PubMed |
Boccalandro HE, Rugnone ML, Moreno JE, Ploschuk EL, Serna L, Yanovsky MJ, Casal JJ (2009) Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis. Plant Physiology 150, 1083–1092.
| Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXnsleiu74%3D&md5=6c158de8e919e662b1f6a6c1640ff4a4CAS | 19363093PubMed |
Cao ZL, Yu QB, Sun Y, Lu Y, Cui YL, Yang ZN (2011) A point mutation in the pentatricopeptide repeat motif of the AtECB2 protein causes delayed chloroplast development. Journal of Integrative Plant Biology 53, 258–269.
| A point mutation in the pentatricopeptide repeat motif of the AtECB2 protein causes delayed chloroplast development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXmtVKit7w%3D&md5=011a3c9a41b5c8e8807667e4e9a46e70CAS | 21294841PubMed |
Casazza AP, Rossini S, Rosso MG, Soave C (2005) Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Molecular Biology 58, 41–51.
| Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmt1yguro%3D&md5=a254c4735f13511422ae861f1d08d079CAS | 16028115PubMed |
Castillon A, Shen H, Huq E (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends in Plant Science 12, 514–521.
| Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXht1KjsbrF&md5=c6647704112d822bba48cc888e4c112aCAS | 17933576PubMed |
Chen M, Tao Y, Lim J, Shaw A, Chory J (2005) Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Current Biology 15, 637–642.
| Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjtV2isbk%3D&md5=520d96c63ebd88350488db68cf3ccdbeCAS | 15823535PubMed |
Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. The Plant Cell 1, 867–880.
| Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3cXkslOj&md5=cb2f805b6c42de09d3c3cdeae3799bcaCAS | 12359912PubMed |
Dalal VK, Tripathy BC (2012) Modulation of chlorophyll biosynthesis by water stress in rice seedlings during chloroplast biogenesis. Plant, Cell & Environment 35, 1685–1703.
| Modulation of chlorophyll biosynthesis by water stress in rice seedlings during chloroplast biogenesis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFKlsrrN&md5=31e71b8f554f4212835be1266273677cCAS |
Dutta S, Mohanty S, Tripathy BC (2009) Role of temperature stress on chloroplast biogenesis and protein import in pea. Plant Physiology 150, 1050–1061.
| Role of temperature stress on chloroplast biogenesis and protein import in pea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXnsleiurk%3D&md5=16e9d2cddf539d2bf0b6cf1a8e790cc2CAS | 19403728PubMed |
Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30, 207–210.
Eisenhart C (1947) The assumptions underlying the analysis of variance. Biometrics 3, 1–21.
| The assumptions underlying the analysis of variance.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaH2s%2FgtF2hug%3D%3D&md5=3d09ebfbf63ac63fcea91f1d66e56b4eCAS | 20240414PubMed |
Fernandez AP, Gil P, Valkai I, Nagy F, Schafer E (2005) Analysis of the function of the photoreceptors phytochrome B and phytochrome D in Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant & Cell Physiology 46, 790–796.
| Analysis of the function of the photoreceptors phytochrome B and phytochrome D in Nicotiana plumbaginifolia and Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXkvVWqu74%3D&md5=7f875fb6c287825d6d0c149e624ae3b1CAS |
Gordon MJ, Carmody M, Albrecht V, Pogson B (2012) Systemic and local responses to repeated HL stress-induced retrograde signaling in Arabidopsis. Frontiers in Plant Science 3, 303
Haslam TM, Kunst L (2013) Extending the story of very-long-chain fatty acid elongation. Plant Science 210, 93–107.
| Extending the story of very-long-chain fatty acid elongation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtV2rtbfL&md5=24b462f144a0dfbef23600386512dc45CAS | 23849117PubMed |
Heddad M, Noren H, Reiser V, Dunaeva M, Andersson B, Adamska I (2006) Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiology 142, 75–87.
| Differential expression and localization of early light-induced proteins in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XpvVKiu7Y%3D&md5=deaf63fe4976e1242ab405fb9b6b8612CAS | 16829586PubMed |
Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305, 1937–1941.
| Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXnslSmu7s%3D&md5=14d2c1020a75663ba66a6a0729ecda5aCAS | 15448264PubMed |
Kessler F, Schnell D (2009) Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Current Opinion in Cell Biology 21, 494–500.
| Chloroplast biogenesis: diversity and regulation of the protein import apparatus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXps1CltLY%3D&md5=7514be0c2486d3a07bce255426426597CAS | 19410443PubMed |
Kim C, Lee KP, Baruah A, Nater M, Gobel C, Feussner I, Apel K (2009) 1O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid. Proceedings of the National Academy of Sciences of the United States of America 106, 9920–9924.
| 1O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXotFaisLs%3D&md5=fa770c9efbc4082abbef2ae7412c07c7CAS | 19482940PubMed |
Krall L, Reed JW (2000) The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proceedings of the National Academy of Sciences of the United States of America 97, 8169–8174.
| The histidine kinase-related domain participates in phytochrome B function but is dispensable.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXkvFCmsLc%3D&md5=da15ccd3cc349921c9fc27c321a06c20CAS | 10869441PubMed |
Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 104, 10270–10275.
| EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmvVCmsL4%3D&md5=0750be22d648458a961d3ac1da9dfd3aCAS | 17540731PubMed |
Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH (2008) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. The Plant Cell 20, 337–352.
| The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXkslSitbw%3D&md5=23191a089c75a6251f8edee9366617f6CAS | 18252845PubMed |
Lucyshyn D, Wigge PA (2009) Plant development: PIF4 integrates diverse environmental signals. Current Biology 19, R265–R266.
| Plant development: PIF4 integrates diverse environmental signals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXjsFWru7c%3D&md5=2f5c40b2868c6fa5bcfc2a4825d02409CAS | 19321147PubMed |
Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. The Plant Cell 15, 1480–1495.
| HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXkvVekt74%3D&md5=0147d9e2e407102f388aa08204021e4aCAS | 12782738PubMed |
Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 98, 12826–12831.
| FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXotFahsLo%3D&md5=5621b039093290d1c3d33f29d8dac5e0CAS | 11606728PubMed |
Mira-Rodado V, Sweere U, Grefen C, Kunkel T, Fejes E, Nagy F, Schafer E, Harter K (2007) Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis. Journal of Experimental Botany 58, 2595–2607.
| Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXpvFWitbs%3D&md5=e8ae9d68f7b4ed5306c677f4f30acb8bCAS | 17545225PubMed |
Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W (2007) The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. The Plant Cell 19, 1313–1328.
| The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmsFeltL0%3D&md5=f082352b8874a698d9b240b83aa6f7b5CAS | 17416734PubMed |
Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proceedings of the National Academy of Sciences of the United States of America 101, 16091–16098.
| The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVWks7rI&md5=374c0660087320fdffae270f63fcebdfCAS | 15505214PubMed |
Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 105, 9433–9438.
| PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXosFartr8%3D&md5=30ebc48304e47413a908c4fbbd57104cCAS | 18591656PubMed |
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497.
Narsai R, Howell KA, Millar AH, O’Toole N, Small I, Whelan J (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. The Plant Cell 19, 3418–3436.
| Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXns1entQ%3D%3D&md5=2e03b991371b6d7d90e37434cd716d0cCAS | 18024567PubMed |
op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, Nater M, Apel K (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. The Plant Cell 15, 2320–2332.
| Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXotlGmtLo%3D&md5=d3f9c9c9cf0ecdafaf8027ca68341d32CAS | 14508004PubMed |
Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiology 155, 1545–1551.
| Genetic dissection of chloroplast biogenesis and development: an overview.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXkvVOrsLk%3D&md5=6cda8dd77b107c4e80553c67b2f57846CAS | 21330494PubMed |
Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. The Plant Cell 5, 147–157.
| Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXks1yhsL4%3D&md5=0f0f948a5c33394940d5bd33e7e2dc7cCAS | 8453299PubMed |
Rose JK, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant & Cell Physiology 43, 1421–1435.
| The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXhtF2gsw%3D%3D&md5=367fe38018727270f1861a19afbbdb03CAS |
Ross J, Li Y, Lim E, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biology 2, S3004
Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, Howell KA, Whelan J, Kazan K, Pogson BJ (2007) Systemic and intracellular responses to photooxidative stress in Arabidopsis. The Plant Cell 19, 4091–4110.
| Systemic and intracellular responses to photooxidative stress in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhvF2htL0%3D&md5=4255874e0ffcd79a0491cfe50e9ca85aCAS | 18156220PubMed |
Sakamoto W, Miyagishima SY, Jarvis P (2008) Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. The Arabidopsis Book 6, e0110
| Chloroplast biogenesis: control of plastid development, protein import, division and inheritance.Crossref | GoogleScholarGoogle Scholar | 22303235PubMed |
Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proceedings of the National Academy of Sciences of the United States of America 106, 7660–7665.
| Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmt1KqtLw%3D&md5=fec21367ca33f28a3245858a8ab08b18CAS | 19380720PubMed |
Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proceedings of the National Academy of Sciences of the United States of America 106, 7654–7659.
| PIF3 is a repressor of chloroplast development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmt1Kqt7w%3D&md5=932b78149de5c7b68a06012d237201a2CAS | 19380736PubMed |
Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Developmental Biology 260, 289–297.
| From seed to seed: the role of photoreceptors in Arabidopsis development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXmt1Wqt7o%3D&md5=a321244d39ad6b1177fa02ccc71d666eCAS | 12921732PubMed |
Sun CW, Huang YC, Chang HY (2009) CIA2 coordinately up-regulates protein import and synthesis in leaf chloroplasts. Plant Physiology 150, 879–888.
| CIA2 coordinately up-regulates protein import and synthesis in leaf chloroplasts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXnsleitbo%3D&md5=6ef1876e1d7748861e5b55a9ae4e8e89CAS | 19386807PubMed |
Swiezewski S, Crevillen P, Liu F, Ecker JR, Jerzmanowski A, Dean C (2007) Small RNA-mediated chromatin silencing directed to the 3ʹ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proceedings of the National Academy of Sciences of the United States of America 104, 3633–3638.
| Small RNA-mediated chromatin silencing directed to the 3ʹ region of the Arabidopsis gene encoding the developmental regulator, FLC.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXjtVWrsrY%3D&md5=92d31609308d5abcf074e6036e873cadCAS | 17360694PubMed |
Tanz SK, Kilian J, Johnsson C, Apel K, Small I, Harter K, Wanke D, Pogson B, Albrecht V (2012) The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LCHB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. The Plant Journal 69, 743–754.
| The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LCHB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XktFWjsbs%3D&md5=e4756f31c9729a27737e54adbf01bf70CAS | 22040291PubMed |
Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. The Plant Cell 21, 1109–1128.
| GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXntFams7o%3D&md5=0e62f4e174fab72ef6e0177af0b481a8CAS | 19376934PubMed |