From little things big things grow: karrikins and new directions in plant development
Mark T. WatersSchool of Molecular Sciences and ARC Centre of Excellence in Plant Energy Biology, The University of Western Australia, Perth, WA 6009, Australia. Email: mark.waters@uwa.edu.au
Functional Plant Biology 44(4) 373-385 https://doi.org/10.1071/FP16405
Submitted: 13 November 2016 Accepted: 16 January 2017 Published: 28 February 2017
Abstract
Karrikins are a family of compounds generated via the incomplete combustion of plant matter. Since their discovery as seed germination stimulants in 2004, a great deal has been learned about the chemistry and the biological mode of action of karrikins. Much interest and progress have stemmed from the structural similarity of karrikins to that of strigolactones – the shoot branching hormone. This review will provide a historical account of some of the more significant discoveries in this area of plant biology. It will discuss how the study of these abiotic signalling molecules, combined with advances in our understanding of strigolactones, has led us towards the discovery of new mechanisms that regulate plant growth and development.
Additional keywords: bioassay, chemical biology, plant development, plant hormone, signalling, strigolactone.
Karrikins: chemical germination cues from fire
The evolution of oxygenic photosynthesis and the subsequent emergence of land plants provided the necessary conditions for fire on Earth. Deposits of fossil charcoal indicate that wildfires began in the Silurian some 420 million years ago, and varied in prevalence according to atmospheric oxygen levels and shifts in dominant terrestrial vegetation (Scott and Glasspool 2006). As an ancient and widespread phenomenon, fire has served as a pronounced evolutionary force that has shaped the composition of ecosystems across the globe (Pausas and Keeley 2009). The passage of a fire event presents several opportunities for regeneration as a result of reduced competition for resources. Accordingly, a large number of plant species have evolved the ability to perceive and respond to fire events. A prominent adaptation among species from fire-prone regions is the promotion of seed germination following exposure to smoke, and chemicals therein (Van Staden et al. 2000; Nelson et al. 2012).
Clues about the chemical nature of smoke compounds with the capacity to stimulate seed germination came from early experiments with ‘liquid smoke’ or ‘smokewater’, a complex solution of combustion products generated by bubbling smoke through water (Baldwin et al. 1994; Baxter et al. 1995; van Staden et al. 1995). The presence of active compounds in smoke indicated that they were at least partially volatile, and chromatography-based fractionation experiments revealed that several different compounds are active, depending on the starting material (Baldwin et al. 1994; van Staden et al. 1995). Despite these promising advances, discovery of an active compound was not made until 2004, using bioassay-guided fractionation of smokewater, followed by gas chromatography-mass spectrometry and nuclear magnetic resonance techniques for structural elucidation. The first report of the butenolide 3-methyl-2H-furo[2,3-c]pyran-2-one was made by chemists at the University of Western Australia using burnt cellulose as a source (Flematti et al. 2004). A subsequent report of the same compound was made by a South African team using a similar approach but with burnt plant material (van Staden et al. 2004). This compound (Fig. 1a) was eventually named ‘karrikinolide’ or KAR1 (after the word ‘karrik’, meaning smoke in the language of the Noongar people indigenous to the Australian south-west). Subsequent work identified several synthetic KAR1 analogues with germination-promoting activity, all containing a consistent butenolide moiety but with varying substituents (Flematti et al. 2007). Crucially, five of these analogues were also detected at varying levels in smokewater, suggesting that they may each contribute to the bioactivity of smoke and char from burned vegetation (Flematti et al. 2009). In keeping with nomenclature for other groups of plant growth regulators (e.g. cytokinins, gibberellins), KAR1 and its analogues KAR2 to KAR6 are collectively known as karrikins (Fig. 1a).

|
It is noteworthy that although karrikins were initially isolated from smoke, most of the KAR1 produced during combustion is retained within the char residue, and KAR1 is sparingly soluble in water (Flematti et al. 2008). As such, the stimulatory activity of karrikins may not disperse very far from the site of a fire. Furthermore, other bioactive germination stimulants besides karrikins are produced during fires, which may have differing physicochemical properties. These include the cyanohydrin glyceronitrile, which slowly hydrolyses to produce cyanide, and which in turn promotes germination in some smoke-responsive species that are otherwise insensitive to karrikins (Flematti et al. 2011).
Seed germination responses to karrikins
Karrikins were isolated by germination-based bioassays using, primarily, seed of lettuce (Lactuca sativa cv. Grand Rapids, Asteraceae). This species is exceptionally sensitive to KAR1, responding to concentrations of 1 nM or below (Flematti et al. 2004). Additional bioassay species included Solanum orbiculatum (Solanaceae; ~10 nM) and Emmenanthe penduliflora (Boraginaceae; ~10 nM) (Flematti et al. 2007). Since the identification of KAR1, several diverse species have been classified as responding positively to karrikins. These include the arable weeds Brassica tournefortii and Sisymbrium orientale (Brassicacaeae) (Stevens et al. 2007), and Avena fatua and Sorghum halepense (Poaceae) (Daws et al. 2007). A recent study of the effects of smokewater and KAR1 on seed germination of 13 species from South China identified only one (Aristolochia debilis; Aristolochiaceae) as responding positively to 1 nM KAR1 or more (Zhou et al. 2014). However, classifying a species in a binary fashion as karrikin-responsive or karrikin-non-responsive is fraught with difficulties, because environmental factors (e.g. maternal life history and wet–dry cycling regimes) that influence physiological seed dormancy in turn affect the capacity of seed to respond to karrikins (Long et al. 2010, 2011). Indeed, varying degrees of physiological seed dormancy between batches of seed from the same species can lead to apparently opposite responses to karrikins (Stevens et al. 2007). It is also possible that ‘non-responsive’ species have incompatible perception machinery, perhaps because, as discussed below, the relevant receptor protein has undergone selection to detect compounds other than karrikins. Accordingly, it is difficult to estimate the number of karrikin-responsive species based on germination tests alone. However, it is clear that the capacity to respond to smoke, and to karrikins specifically, is taxonomically widespread among angiosperms.
For the few species tested, KAR1 is the generally the most effective germination stimulant among the karrikins, which makes evolutionary sense in light of KAR1 being the most abundant karrikin produced by combustion of cellulose. As a germination stimulant, KAR1 is ~10-fold more effective than KAR2 in Grand Rapids lettuce, and 100-fold more effective in S. orbiculatum (Flematti et al. 2007). All else being equal, karrikins with a methyl group at the C-3 position on the butenolide ring are generally more active than those without (Flematti et al. 2007). Few studies have explored the sensitivity of different species to the various karrikin family members, so it is not yet clear how widespread this pattern is. Nevertheless, a notable reversal of the preference for KAR1 over KAR2 is found in Arabidopsis thaliana, as discussed below.
Other plant responses to karrikins
Karrikins were discovered on the basis of their ability to promote seed germination and overcome seed dormancy, but they are widely reported also to enhance seedling survival and seedling vigour. Both smokewater (1 : 500 dilution) and KAR1 (100 nM) led to increased shoot and root growth in tomato, okra, maize and rice (Kulkarni et al. 2006; van Staden et al. 2006). In maize, a single 1 h soak of kernels in 100 nM KAR1 enhanced post-germination seedling growth relative to untreated controls when measured 30 days after germination (although effects at earlier or later growth stages were not reported). The same treatment also led to significantly improved seedling survival rates (van Staden et al. 2006). KAR1 was also shown to improve germination of tomato seed and subsequent seedling vigour when grown at low (10°C) or high (40°C) temperatures (Jain et al. 2006). These findings suggest that karrikins might be powerful agents for seed preconditioning to improve crop productivity under stressful conditions.
Recently, it was demonstrated that biochars – charcoal-like material produced by pyrolysis of organic material under oxygen-limiting conditions – contain readily detectable amounts of KAR1 (Kochanek et al. 2016). The yield of KAR1 varied greatly depending on the pyrolysis technology used, but reached 82 ng per 100 g (~1 ppb) of biochar. Liquid by-products of one biochar process contained over 400 nM KAR1, which compares favourably with smokewater (Fig. 1a). Pertinently, the shoot length of both tomato and lettuce plants grown in soil containing up to 10% biochar was significantly greater than untreated controls after two weeks of growth (Kochanek et al. 2016). In addition, the KAR1 content of biochar correlated positively with the effects on plant growth, tempting one to conclude that karrikins are responsible for the benefits of biochars. However, the physiological changes that karrikins and biochar bring about to stimulate plant growth remain to be investigated in any detail, and such studies would benefit greatly from the inclusion of genetic resources.
Karrikin studies in Arabidopsis
A great deal of our knowledge on the effects and mechanisms of karrikins on plant growth come from studies on the genetic workhorse A. thaliana. The seed of this species exhibits variable degrees of primary seed dormancy, defined as the tendency for freshly harvested seed not to germinate under otherwise favourable conditions. The depth of primary dormancy depends heavily on the ecotype in question: Cape Verde Islands (Cvi-0) is highly dormant, whereas Landsberg erecta (Ler) is less so (Alonso-Blanco et al. 2003). Columbia-0 (Col-0) meanwhile exhibits little primary dormancy at all (van der Schaar et al. 1997). Therefore, as with other species, applying karrikins to stimulate seed germination has variable effect on the ecotypes of Arabidopsis. Nevertheless, primary dormant Ler seeds demonstrate a robust response to 10 nM KAR2, and to 100 nM KAR1 and KAR3 (Nelson et al. 2009). Further experiments showed that KAR1 cannot overcome the requirement of germination for gibberellin (GA) biosynthesis, and indeed that KAR1 might act in part by stimulating the expression of the GA biosynthetic genes GA3ox1 and GA3ox2, which would, in turn, stimulate germination (Nelson et al. 2009).
The developmental response of a seedling to light is known as photomorphogenesis. In epigeal seedlings like Arabidopsis – where the cotyledons emerge above ground and become photosynthetic – photomorphogenesis is characterised by greening and expansion of the cotyledons, reduction or cessation of hypocotyl elongation, and increased root growth relative to dark-grown seedlings. Treatment of wild-type Arabidopsis seedlings with 100 nM KAR1 or KAR2 led to an enhancement of these traits, especially cotyledon expansion and inhibition of hypocotyl elongation (Nelson et al. 2010). As with seed germination, Arabidopsis seedlings were more responsive to KAR2 than to KAR1. Karrikins also reduced the minimum amount of red light required to trigger germination after an inhibitory far-red light pulse; however, karrikins were insufficient to overcome the requirement for light altogether, suggesting that they act downstream of phytochrome B in the control of germination and photomorphogenesis (Nelson et al. 2010). Nevertheless, karrikins do induce transcriptional changes in darkness, suggesting that some karrikin responses are light-independent (Waters and Smith 2013). Together, these findings indicate that karrikins enhance the sensitivity of seedlings and seed to light, and act as general positive regulators of light-dependent development.
As a northern hemisphere temperate species with no ecological requirement for fire regimes, the response of A. thaliana to karrikins was a critical finding. First, it provided a powerful means to investigate, using genetics, the molecular basis for the perception and response of plants to karrikins. In addition, it indicated that these mechanisms are likely to be conserved among angiosperms, aiding the transfer of knowledge gleaned from one species to others of ecological or economic significance. Most importantly, the evolutionary conservation of a trait that is presumably not under selection indicates that the mechanisms for karrikin perception and response may have other functions in plant development beyond the detection of compounds released by fire.
Karrikins are structurally related to strigolactones
Like karrikins, strigolactones are also butenolide compounds (Fig. 1b), and the structural similarity between these compounds was quickly recognised (Flematti et al. 2004). Strigolactones were first discovered in the 1960s and ’70s as compounds exuded from plant roots that induced the germination of seed of root parasitic weeds in the Orobanchaceae (reviewed by Ruyter-Spira et al. 2013). But why should a plant synthesise and secrete compounds that might cause self-harm through parasitism? In 2005, Akiyama et al. (2005) demonstrated that natural strigolactones (such as 5-deoxystrigol), and the synthetic strigolactone analogue GR24, could stimulate germination and hyphal branching in fungal spores that form arbuscular mycorrhizal (AM) symbioses. These associations, in which the fungi assist the plant in mineral uptake in exchange for organic carbon, are ubiquitous throughout the plant kingdom, even though a few families (such as the Brassicaceae) have lost the ability to form them (Bouwmeester et al. 2007; Bravo et al. 2016). Therefore strigolactones were shown to constitute the elusive ‘branching factor’ that enabled host plants to recruit AM fungi, especially under nutrient-limiting conditions. The biosynthetic source of strigolactones was unclear, but carotenoids were considered a likely possibility based on inhibitor studies (Matusova et al. 2005).
In parallel, plant developmental biologists and geneticists were seeking to understand the mechanistic basis for controlling shoot architecture – specifically the regulation of shoot branching. Several studies of mutants with increased shoot branching phenotypes in pea (Pisum sativum), rice (Oryza sativa), petunia (Petunia hybrida) and Arabidopsis had indicated that a mobile substance, synthesised in and/or transported from the roots, could inhibit branching in the shoot and thus promote apical dominance (Beveridge et al. 1996; Napoli 1996; Beveridge et al. 1997; Stirnberg et al. 2002; Ishikawa et al. 2005). Two of the affected genes in these mutants were found to encode carotenoid cleavage dioxygenase (CCD) enzymes, thus implicating carotenoids as a possible source (Sorefan et al. 2003; Booker et al. 2004; Johnson et al. 2006; Arite et al. 2007; Drummond et al. 2009). In 2008, these lines of evidence were brought together to show that strigolactones were endogenous plant hormones responsible for controlling shoot architecture (Gomez-Roldan et al. 2008; Umehara et al. 2008). Since this major step forward was made, strigolactones have been implicated in numerous other processes, including leaf senescence (Snowden et al. 2005; Yamada et al. 2014), adventitious rooting (Rasmussen et al. 2012), secondary thickening (Agusti et al. 2011), and regulation of root architecture under changing nutrient conditions (Ruyter-Spira et al. 2011; Mayzlish-Gati et al. 2012; reviewed by Al-Babili and Bouwmeester 2015; Waters et al. 2017).
At the time of the discovery of karrikins in 2004, little more about the function of strigolactones was known, and the significance of the structural similarities was not clear. As both were butenolide germination stimulants, it was reasonably hypothesised that karrikins might be abiotic mimics of strigolactones with similar modes of action (Flematti et al. 2004). However, it was soon shown that the relationship was not so straightforward. First, although karrikins were potent germination stimulants of Brassica tournefortii, the synthetic strigolactone GR24 was not (Nelson et al. 2009). Conversely, seed of the parasitic weed Orobanche minor responded strongly to GR24, but not at all to karrikins; this observation has since been extended to several other parasitic species (Conn et al. 2015). These observations may simply reflect species-specific adaptation towards different germination stimulants. However, karrikins were also inactive as inhibitors of shoot branching, unlike GR24 (Nelson et al. 2011). Thus, physiological evidence was building that karrikins and strigolactones have distinct bioactivities and, therefore, different perception mechanisms. More direct molecular–genetic evidence has subsequently solidified this position, but we now know that the parallels between karrikins and strigolactones extend far beyond their superficially similar chemical structures.
Genetic analysis of karrikin perception and signalling
2011: MAX2
Genetic screens for Arabidopsis mutants deficient in karrikin signalling have been central to building our knowledge of the function of karrikins. The first screen for karrikin insensitive (kai) mutants involved the selection of mutagenised, primary dormant seed that failed to germinate on water-agar medium supplemented with karrikins. This screen identified two allelic mutants called kai1, which, besides impaired seed and seedling responses to karrikins, also exhibited several other phenotypes (Nelson et al. 2011). These included abnormal seedling development, curled, rounded leaves, increased shoot branching, and reduced stature. These phenotypes are also characteristic of the strigolactone-insensitive mutant more axillary branches2 (max2) (Stirnberg et al. 2002). This insight led Nelson et al. (2011) to make an educated guess, and upon sequencing they found that both kai1 alleles contained frameshift mutations in MAX2. This was a spectacular discovery because not only did it demonstrate that MAX2 had a new role in seed germination, but also it showed that the mechanism for the perception of strigolactones and karrikins shared a common component (Waters et al. 2011).
MAX2 is an F-box protein, a family of leucine-rich-repeat proteins that form one part of the SCF class of E3 ubiquitin ligase complexes (Xu et al. 2009). The F-box component confers substrate specificity to polyubiquitination, a process that labels target proteins for proteolytic destruction via the 26S proteasome. An SCF E3 ubiquitin ligase that contains MAX2 can be denoted SCFMAX2. The perception mechanisms for several plant hormones, including auxins, gibberellins and jasmonates make use of F-box proteins to selectively target downstream repressor proteins (McGinnis et al. 2003; Dharmasiri et al. 2005; Kepinski and Leyser 2005; Katsir et al. 2008). Thus, following the identification of MAX2, additional karrikin signalling and response components were anticipated.
2012: KAI2/HTL and D14
MAX2 (and its homologue in rice, DWARF3) was known to be essential for strigolactone perception. As the auxin receptor, TIR1, was also an F-box protein, it was possible that MAX2 also served as a receptor strigolactones and karrikins. However, there was no molecular evidence for this hypothesis, and no obvious mechanism for how MAX2 might mediate such different physiological responses to the two different classes of compounds. In 2009, the rice mutant dwarf14 (d14; also known as d88 or htd2) was described as having a shoot architecture phenotype largely indistinguishable from d3 (Arite et al. 2009; Liu et al. 2009). Crucially, like d3, d14 was also strigolactone-insensitive, and double mutant analysis placed it within the strigolactone response pathway (Arite et al. 2009). DWARF14 encodes an α/β-fold hydrolase, and its function was not described until 2012, but it was known to be part of a larger family of proteins common to all land plants. Phylogenetic analysis indicated that the Arabidopsis and rice genomes each contained a single D14 orthologue, as well as a more distant paralogue dubbed D14-LIKE. At the time, this gene also had no known function, with no described mutant phenotype in any species. As the gibberellin receptor GID1 is an α/β-fold hydrolase (Ueguchi-Tanaka et al. 2005), the possibility that D14 might serve as the strigolactone receptor was an appealing hypothesis. D14 and D14-LIKE thus represented potentially valuable signalling components that remained to be investigated.
On the supposition that D14 and D14-LIKE proteins might be receptor proteins, and in turn might provide MAX2 with a means to discriminate between karrikins and strigolactones, Waters et al. (2012a) set out to analyse the function of D14 family members in Arabidopsis. As expected, the d14 mutant of Arabidopsis exhibited an increased shoot branching phenotype and strigolactone insensitivity (Waters et al. 2012a; Chevalier et al. 2014). By a remarkable stroke of serendipity, we converged upon the function of D14-LIKE by two parallel routes. First, as part of a systematic search for karrikin insensitive mutants among candidate genes known to be involved in seed germination and dormancy, we discovered a mutant allele of the gene SPATULA (At4g36930). Although this allele, spt-1, was almost completely insensitive to karrikins in seed germination and seedling hypocotyl elongation assays, several other spt alleles (including predicted null alleles) responded normally to karrikins. Eventually we were forced to conclude that a second, unknown mutation elsewhere in the spt-1 genome was responsible for the karrikin insensitive phenotype, and we renamed this mutant karrikin insensitive2–1 (kai2–1) after backcrossing to segregate away the spt-1 mutation. Recombinant F2 individuals were much less frequent than expected assuming independent assortment, suggesting that the two mutations were closely linked.
Meanwhile, in the second route, we used reverse genetics to isolate a Ds transposon insertion mutant in the Arabidopsis orthologue of rice D14-LIKE, At4g37470. We soon noticed that SPATULA (At4g36930) and At4g37470 were closely located on the same chromosome, prompting us to sequence At4g37470 in the kai2–1 mutant background. In doing so, we discovered a G-to-A transition in the coding sequence, modifying a conserved glycine residue to glutamic acid (Waters et al. 2012a). Coupled with allelism tests, this finding indicated that At4g37470 was in fact KAI2, and we renamed the Ds insertion allele kai2–2. In the course of our work, another group identified a further kai2 allele in a screen for photomorphogenesis mutants (Sun and Ni 2011). They named the mutant hyposensitive to light (htl) on the basis of its elongated hypocotyl phenotype. We felt that KAI2 was a more accurate description of the function of the gene, but both names are now widely adopted in the literature (e.g. Toh et al. 2015), as is D14-LIKE in studies on rice (e.g. Kameoka and Kyozuka 2015).
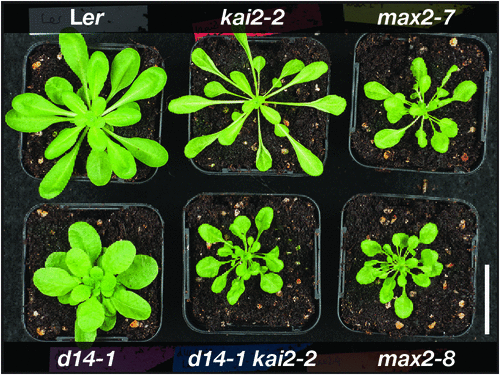
A key observation from isolating the Arabidopsis d14 and kai2 mutants was that each one exhibited different components of the max2 phenotype. Although d14 seedlings were phenotypically normal, kai2 seedlings were indistinguishable from max2 seedlings. Meanwhile, d14 mutants exhibited the same increased shoot branching phenotype as max2, but shoot branching was normal in kai2. Thus, the max2 phenotype could be considered to be an amalgamation of kai2 and d14 phenotypes; accordingly, a kai2 d14 double mutant fully recapitulates the max2 phenotype (Fig. 2). Importantly, kai2 mutants retain the ability to respond to exogenous strigolactones, and d14 mutants to karrikins (Waters et al. 2012a; Scaffidi et al. 2014). By extension, some max2 phenotypes are thus strigolactone related (as mediated by D14), and others are strigolactone-independent (as mediated by KAI2).
|
KAI2 and D14 are capable of mediating plant responses to very similar compounds. Although KAI2 is essential for karrikin responses, in Arabidopsis it can also detect the synthetic strigolactone GR24 (Waters et al. 2012a). At first glance, this result suggests that KAI2 can serve as a receptor for natural, endogenous strigolactones; however, in Arabidopsis at least, this is not the case. Careful mutant analysis indicated that the KAI2-dependent activity of GR24 is an artefact of the racemic composition of standard preparations of synthetic GR24 (Scaffidi et al. 2014). Although all natural strigolactones contain a 2ʹR-configured D-ring, racemic GR24 (rac-GR24) contains an equal proportion of 2ʹR and 2ʹS stereoisomers (Fig. 1). These mirror images, or enantiomers, have been given the simplified names of GR245DS and GR24ent–5DS, respectively, in reference to the natural strigolactone 5-deoxystrigol (5DS) that they most closely resemble (Scaffidi et al. 2014; Flematti et al. 2016). GR24ent–5DS can substitute for karrikins in a range of KAI2-dependent assays, albeit with reduced sensitivity (Scaffidi et al. 2014; Waters et al. 2015a, 2015b). Likewise, D14 responds preferentially to GR245DS over GR24ent–5DS (Nakamura et al. 2013; Scaffidi et al. 2014; Waters et al. 2015b; Zhao et al. 2015). The biological significance of KAI2 recognising non-naturally-configured strigolactones is unclear, but it does demonstrate that KAI2 and D14 exhibit distinct substrate preferences, which in part explains their functional differences. In Arabidopsis at least, KAI2 does not appear to have any role in mediating responses to endogenous strigolactones.
Several angiosperms, including Arabidopsis and rice, contain a third member of the D14 family. This protein – D14-LIKE2 (DLK2) – is a close paralogue of D14, suggesting that it arose through a duplication of D14 before the divergence of monocots and dicots. At the time of writing, the function of DLK2 remains an enigma, but a very curious one. DLK2 transcripts are strongly upregulated in response to application of karrikins, making it a convenient transcriptional marker for KAI2-dependent activity (Waters et al. 2012a). DLK2 transcripts are also repressed in kai2 and max2 seedlings, but not in d14 seedlings; however, D14-mediated signalling of exogenous strigolactones can increase the abundance of DLK2 transcripts (Waters et al. 2012a; Scaffidi et al. 2014). These observations could suggest that DLK2 has some form of negative feedback role, perhaps serving to repress KAI2- or D14-dependent signalling. However, dlk2 mutants have no obvious seed germination or seedling development phenotype, and shoot branching is apparently normal (Waters et al. 2012a). A closer examination of dlk2 phenotypes, perhaps in conjunction with higher order mutants, will doubtless reveal DLK2 function in time.
2013: The SMXL family
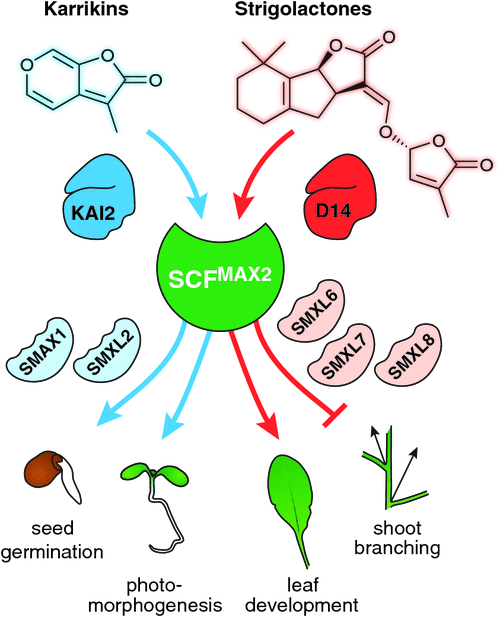
The newly identified function for MAX2 in karrikin signalling presented a new angle to discover the putative repressor proteins that are targeted for proteolysis by SCFMAX2. A suppressor screen for the karrikin-related max2 phenotypes relating to seed germination and seedling growth identified SUPPRESSOR OF MAX2 1 (SMAX1) (Stanga et al. 2013). Arabidopsis seedlings deficient in SMAX1 and, to a lesser extent its close paralogue SMAX1-LIKE 2 (SMXL2), resemble wild type seedlings treated with karrikins – i.e. they exhibit constitutive karrikin responses (Stanga et al. 2016). Shortly after the discovery of SMAX1, the rice protein DWARF53 (D53) was identified thanks to a dominant rice mutant that had similar phenotypes to d3 and d14, such as high tillering and increased strigolactone production (Jiang et al. 2013; Zhou et al. 2013). D53 is another member of the SMXL family, all of which share similarity to the ClpB/HEAT SHOCK PROTEIN 100 (HSP100) class of heat shock proteins. In Arabidopsis, three D53 homologues (SMXL6, SMXL7 and SMXL8) act redundantly to mediate all tested strigolactone-related components of MAX2 function (Soundappan et al. 2015; Liang et al. 2016; Bennett et al. 2016). This redundancy might explain why suppressor screens for the increased shoot branching phenotype of max2 had not uncovered this protein family. Instead, one such screen identified a role for the transcription factor FAR RED ELONGATED HYPOCOTYL3 in the regulation of auxin-dependent axillary bud outgrowth (Stirnberg et al. 2012). The functions of other SMXL members – SMXL3, SMXL4 and SMXL5 in Arabidopsis – are yet to be resolved (Khosla and Nelson 2016). However, it is clear that the diverse functions of MAX2 relating to karrikins and strigolactones are a combinatorial result of two different receptor proteins (KAI2 and D14), and two subclasses of downstream repressor proteins (SMAX1/SMAXL2, and SMXL6/7/8) (Fig. 3). A significant body of work has resulted in a molecular model for strigolactone and karrikin signalling mechanisms, and this has been reviewed recently (Waters et al. 2017).
|
KAI2 and D14 are both receptor proteins and enzymes
D14 and KAI2 are unusual among hormone receptor proteins because they are also functional enzymes with hydrolytic activity. In 2012, the structure of the D14 homologue in Petunia hybrida, DAD2, was solved (Hamiaux et al. 2012). Shortly thereafter, structures of KAI2 and D14 from Arabidopsis and rice were also published (Bythell-Douglas et al. 2013; Guo et al. 2013; Kagiyama et al. 2013; Zhao et al. 2013). Both KAI2 and D14 contain a catalytic triad of Ser, His and Asp residues at the end of a hydrophobic ligand-binding cavity. The overall structure is globular, consisting of a core domain of seven α-helices and seven β-sheets linked to a ‘lid’ domain of four α helices. The ligand binding cavity sits between these two domains.
As part of the strigolactone signalling pathway, D14 has received much attention to demonstrate its role as a receptor, a position supported by a substantial body of evidence. As described above, loss-of-function d14 mutants are consistently SL-insensitive in several species (Arite et al. 2009; Waters et al. 2012a; Hamiaux et al. 2012; Chevalier et al. 2014; de Saint Germain et al. 2016). Extensive biochemistry experiments have enhanced this genetic evidence. D14 hydrolyses strigolactones, albeit very slowly, by nucleophilic attack upon the strigolactone D-ring, which is oriented towards the catalytic serine (Hamiaux et al. 2012; Yao et al. 2016; de Saint Germain et al. 2016). Mutation of the catalytic triad residues renders D14 non-functional as an enzyme, and also as a transducer of strigolactone signalling in planta (Hamiaux et al. 2012). A critical observation is that binding of strigolactone to D14 induces a marked change in protein melting temperature, suggesting that the ligand-protein interaction imparts a change in D14 conformation (Hamiaux et al. 2012; Zhao et al. 2013; Abe et al. 2014; Waters et al. 2015b). Consistent with this interpretation, crystallography demonstrated the ligand-dependent interaction of D14 and D3/MAX2, together with the Arabidopsis SCF. component ASK1 (Yao et al. 2016). This crucial evidence revealed that binding of strigolactone to GR24 causes the lid domain of D14 to flatten and partially collapse the ligand-binding cavity, trapping a strigolactone hydrolysis product within. At the same time, another study determined that this hydrolysis product was covalently linked to the D14 catalytic His residue (de Saint Germain et al. 2016). Thus, the significance of the enzymatic activity of D14 has become clearer: substrate hydrolysis is an integral step to form a catalytic intermediate that enhances the formation of a protein complex, which in turn transduces the signal. Hydrolysis might also serve as a mechanism for destroying the strigolactone ligand and deactivating the signal. At least in vitro, the hydrolysis product is slowly released, suggesting that D14 might relax after activation. However, in plants D14 undergoes proteolysis in response to strigolactone application (Chevalier et al. 2014), so this may be the dominant mechanism for removal of D14 in its activated and closed state. Finally, strigolactone hydrolysis by D14 is insufficient to confer a signalling function, because the non-functional d14–5 protein exhibits enhanced rates of hydrolysis, but strongly impaired interaction with D3/MAX2 (Yao et al. 2016). Collectively, these data demonstrate conclusively that D14 is not merely an enzyme, but also serves as a strigolactone receptor that binds and subsequently hydrolyses its ligand to transduce a signal.
Given this abundant knowledge about D14, the case for KAI2 being the karrikin receptor is very strong. KAI2 is an active hydrolase, and mutagenesis of the catalytic triad renders it inactive both in vitro as an enzyme and in planta as a signal transducer (Waters et al. 2015a, 2015b). Several studies have detected direct interaction between KAR1 and KAI2 homologues using isothermal calorimetry and/or fluorescence-based binding assays (Guo et al. 2013; Kagiyama et al. 2013; Xu et al. 2016). Two of these studies have also used crystallography to determine the position of KAR1 in the binding pocket (Guo et al. 2013; Xu et al. 2016). However, each study revealed KAR1 to sit in quite different orientations, perhaps because two different KAI2 proteins were investigated – one from Arabidopsis and one from the parasitic plant Striga hermonthica. Neither structure showed a dramatic change in conformation between the KAR1-bound and KAR1-free states. Curiously, karrikins do not induce any thermal destabilisation upon purified KAI2 in the same way that strigolactones do for D14 (Waters et al. 2015b). Thermal destabilisation of KAI2 can, however, come about through addition of the non-natural enantiomer GR24ent–5DS, which is also a hydrolytic substrate for KAI2, suggesting that mechanistically KAI2 and D14 work in a similar fashion as enzyme–receptors. At this time there is no direct evidence that karrikins are hydrolysed by KAI2, perhaps because karrikins are not expected to be destroyed by nucleophillic attack upon the butenolide ring (Scaffidi et al. 2012; Waters et al. 2012b). Thus some data do not fully support a simple interpretation that KAI2 binds and hydrolyses KAR1 directly, with the same outcomes as when D14 binds strigolactones. It is possible that, without additional protein components (i.e. MAX2 and ASK1) to stabilise any conformational change in KAI2, the natural orientation of KAR1 in the KAI2 cavity will not be detected. Alternatively, karrikins might undergo metabolism in vivo to generate a bioactive ligand that is capable of fully activating KAI2.
There are several additional aspects of karrikin signalling that remain unclear. For example, although a physical interaction between D14 and D53/SMXL7 has been demonstrated (Jiang et al. 2013; Zhou et al. 2013; Wang et al. 2015), the same has not yet been established for KAI2 and SMAX1 or SMXL2, despite very strong genetic evidence that they operate in the same pathway. In addition, KAI2 undergoes degradation via an unknown mechanism as a consequence of karrikin signalling, but this process is independent of MAX2 (Waters et al. 2015a). This is in stark contrast to D14, which is degraded via the proteasome in a process that requires MAX2 (Chevalier et al. 2014). These apparent discrepancies may represent functionally significant points of distinction from the proposed mechanism for strigolactone signalling.
Beyond karrikins: what else does KAI2 do?
The broad evolutionary conservation of KAI2 proteins – from algal relatives of land plants to angiosperms – clearly demonstrates that the primary function of KAI2 is unlikely to be the perception of compounds from fire. Although karrikins were instrumental in the discovery of KAI2 – not least because they are a potent activator of KAI2-dependent signalling, even in non-fire-prone plants – we now appreciate that KAI2 regulates some of the wide number of physiological processes that are dependent on MAX2. These include effects on seed germination and seedling development, as are well established, but effects on leaf development are also evident, at least in Arabidopsis (Waters et al. 2012a, 2015b; Bennett et al. 2016) (Fig. 2). In theory, any MAX2-dependent process that cannot be attributed to D14-dependent signalling (i.e. those processes regulated by strigolactones) might be under the influence of KAI2. A notable example of this was recently uncovered by studying the function of KAI2 in rice.
As noted above, strigolactones enhance the formation of AM symbioses by promoting the germination and hyphal development of fungal spores. Although strigolactone production is thus important for successful AM colonisation, it is not necessary, because strigolactone biosynthesis mutants nevertheless develop functional symbioses (Gomez-Roldan et al. 2008; Kohlen et al. 2012; Kretzschmar et al. 2012). Somewhat confusingly, the F-box protein D3/RMS4/MAX2 was shown to be essential for AM symbioses in both rice (Yoshida et al. 2012) and pea (Foo et al. 2013). However, it became clear that D14 was not responsible for regulating this aspect of MAX2 function, because rice d14 mutants did not show the same AM-deficient phenotype as d3 (Yoshida et al. 2012). These authors also excluded a role for KAI2/D14-LIKE in AM symbiosis, based on the analysis of RNAi knockdown lines that showed no phenotypic defect in this regard. This result was puzzling, as it implied that D3/MAX2 had a function in AM colonisation that was independent of both D14 and KAI2. However, in a classical genetic screen for rice mutants unable to form AM associations, Gutjahr et al. (2015) isolated the hebiba mutant, which showed several defects in the symbiotic process. Genetic mapping indicated that the hebiba phenotype resulted from a genomic deletion of 169 kb on chromosome 3. Complementation with genomic clones from this interval eventually identified the causative gene: KAI2, or D14-LIKE (Gutjahr et al. 2015). With this, order was restored: the AM-deficient phenotype of d3 was shared with d14-like, reinforcing the prevailing notion that the functions of MAX2 are divided between D14 and KAI2. Thus, Gutjahr et al. (2015) demonstrated elegantly a new and critical role for KAI2 in the evolutionarily ancient symbiosis between plant roots and fungi. To date, the requirement for KAI2 in this process has not been demonstrated in other species, but it would be surprising if it were a function unique to rice. It remains to be elucidated exactly how KAI2 mediates AM symbiosis, but based upon the lack of AM development and induction of colonisation marker genes, KAI2 appears to act very early stage of the interaction, perhaps during presymbiotic signalling (Gutjahr et al. 2015). It is tempting to speculate that KAI2 perceives a compound present in the fungus. However, karrikins and extracts from germinating fungal spores do not induce the same transcriptomic changes in rice (Gutjahr et al. 2015). Perhaps the level of the KAI2 ligand, or sensitivity to it, increases upon initial interaction with AM, and these changes are necessary to trigger the developmental changes in the plant. In this way, the signalling pathway defined by KAI2, MAX2 and SMAX1 might serve to create a permissive condition for symbiosis. One prediction of this hypothesis would be that loss-of-function smax1 mutants are highly permissive, or at least would suppress the symbiosis-deficient phenotypes of kai2 and max2. Although no smax1 mutant outside Arabidopsis has yet been described, with modern genome editing tools such experiments in rice or other model systems are becoming increasingly feasible.
KAI2 has another, more specialised role in mediating interactions between plant roots and another organism: the detection of host-derived strigolactones by parasitic weeds. Obligate parasitic weeds such as Striga hermonthica and Phelipanche aegyptiaca are widespread agricultural pests, especially in sub-Saharan Africa, where they infest staples such as sorghum, maize and millet, and cause substantial crop losses (Parker 2009). In many parasitic weed genomes, KAI2 is present in multiple copies, having undergone extensive gene duplication. In contrast, although D14 is also present in these genomes – presumably to mediate endogenous strigolactone signalling within the parasite itself – this gene is almost always present in single copy, and only rarely duplicated (Conn et al. 2015). Some of the KAI2 copies are unique to parasites and are particularly divergent from other angiosperm KAI2 genes, suggesting that they underwent increased rates of evolution via positive selection (Conn et al. 2015). These ‘KAI2d’ genes encode proteins with substantially enlarged ligand binding pockets relative to Arabidopsis KAI2, and structural modelling suggested that these pockets could accommodate a strigolactone molecule. Several KAI2d transgenes from S. hermonthica and P. aegyptiaca were able to confer strigolactone-specific germination responses to Arabidopsis kai2 mutants, which normally are insensitive to these compounds (Conn et al. 2015; Toh et al. 2015). In addition, purified KAI2d proteins from S. hermonthica were able to hydrolyse strigolactones, with varying affinities for different strigolactones (Tsuchiya et al. 2015). Thus, these specialised versions of KAI2 are likely responsible for detection of strigolactones by parasitic weeds. It is noteworthy that the evolutionary target for selection in this case was KAI2, which regulates seed germination, rather than D14, which is the canonical strigolactone receptor. This scenario may reflect the relative ease of changing the ligand specificity of a receptor, rather than modifying tissue expression domains and affinities for partner proteins.
It is evident that KAI2 is sensitive to several compounds that regulate its activity and bring about downstream molecular and physiological changes. If the ancestral role for KAI2 is not to perceive karrikins, which are unusual substances for most plants to encounter, then what might this role be? One hypothesis is that KAI2 is a receptor for an unknown endogenous compound with structural similarity to karrikins, but which is distinct from strigolactones derived from carlactone (Waters et al. 2014; Conn and Nelson 2015). The arguments in favour of this notion are multiple, and mainly stem from evolutionary-genetic analysis and from analogy to strigolactones. First, phylogenetic analysis strongly suggests that a KAI2-like protein was the evolutionary ancestor of D14, implying that the signalling function of KAI2 is fundamental and has been retained throughout the diversification of land plants (Delaux et al. 2012; Waters et al. 2012a, 2015b). Given that D14 was an innovation of the seed plants but strigolactones are found in Charophyte algae, it is likely that KAI2 proteins in early land plants were receptors for strigolactones, and perhaps for other signalling compounds as well (Delaux et al. 2012; Waters et al. 2013; Lopez-Obando et al. 2016). Second, KAI2-dependent signalling requires an intact catalytic triad, and this requirement is evolutionarily conserved between lycophytes and angiosperms (Waters et al. 2015a, 2015b). It seems unlikely that a functional catalytic triad of KAI2 would persist across such long timescales were it not required for the perception of a signalling molecule with a similar mechanism to D14 and strigolactones. Fourth, the appearance of KAI2d homologues in parasitic weeds to detect strigolactones implies that the ancestral KAI2 also had the capacity to respond to a similar ligand. Crucially, these ancestral KAI2 homologues – which show relatively reduced rates of evolution and therefore higher degrees of conservation – have retained a strigolactone-independent function similar to that of KAI2 in Arabidopsis (Conn and Nelson 2015; Conn et al. 2015). Fifth, the fact that KAI2- and D14-dependent signalling both operate via the same family of SMXL repressor proteins also suggests that KAI2 likely modulates MAX2 activity through a similar ligand-dependent mechanism. Finally, kai2 and max2 mutants of Arabidopsis both share seed germination and seedling morphogenesis phenotypes that are opposite to the effects of karrikin treatment. These phenotypes imply that kai2 and max2 are unable to perceive some internal signal that karrikins enhance or mimic (Nelson et al. 2011; Waters et al. 2012a). Accordingly, there is good reason to believe that a hormone-like KAI2 ligand (KL) exists.
Chasing a new hormone
Historical discoveries of classical plant hormones have come about through the identification of an activity, followed by the identification of a source of this activity, and then isolation of the active compound. For example, gibberellins were discovered on the basis of the bakanae (‘foolish seedling’) disease of rice, caused by the fungus Gibberella fujikuroi (reviewed by Mander 2003). Overproduction of gibberellic acid by the fungus (the source) causes stem elongation and chlorosis (the activity). Similarly, ethylene was identified as the causative agent from gas streetlights that caused twisting and thickening of stems in nearby trees (Abeles et al. 1992). The discovery of strigolactones as a plant hormone was somewhat different; the existence of a mobile inhibitor of shoot branching was first predicted on the basis of genetics and physiological experiments. Likewise, the prediction of a KAI2 ligand (KL) comes primarily from inference and analogy, rather than from an observed bioactivity of a compound from a biological source.
Direct evidence for a compound that operates through KAI2 and is derived from plant material requires the development of a bioassay that provides a specific response. Recently, Sun et al. (2016) described the creation of a transgenic Arabidopsis line carrying a luciferase (LUC) coding sequence driven by the DLK2 promoter, which was chosen on the basis that DLK2 transcripts are strongly induced by KAI2-dependent signalling. We showed that this reporter gene was sensitive to karrikins and GR24ent–5DS (the non-natural strigolactone enantiomer) but not to other plant hormones. These responses were abolished in a kai2 mutant background, demonstrating the specificity of the reporter for KAI2-dependent activity. Importantly, we showed that extracts from Arabidopsis leaf material could activate expression of the reporter, suggesting that compounds present in leaf tissue were potential activators of KAI2 signalling (Sun et al. 2016). However, this activity was primarily found in the aqueous fraction of the plant extract, which would not be the case for hydrophobic compounds like karrikins and strigolactones. It is thus possible that the activity detected is not a direct ligand of KAI2, but may be a precursor of KL, or a stimulator of KL biosynthesis. Nevertheless, this work provides strong evidence for the existence of compounds that might act via KAI2, and lays out the groundwork for discovering a potential new hormone.
The challenges associated with identifying KL are formidable. The main difficulty will be the presumably very low abundance of KL in plant tissues. As a point of reference, strigolactones are present on the scale of 10 pg g–1 FW in rice roots (Umehara et al. 2008). With the availability of a reporter assay, it should be possible to screen various sources for richer KL content, but without doubt considerable scaling-up of material will be necessary to obtain quantities sufficient for spectroscopic analysis. In addition, the compound(s) in question might be unstable, especially in aqueous conditions, which will impose difficulties during separation and purification steps. Ongoing development and improvement in bioassays that make full use of our knowledge of the molecular mechanisms of KAI2-dependent signalling, combined with modern synthetic biology approaches, will improve the chance of success.
Concluding remarks
So, what has studying karrikins taught us? Their discovery came about through curiosity-driven investigations into a relatively obscure environmental sensing mechanism: the ability of seeds to detect a recent fire event. We now recognise that karrikins act by hijacking a taxonomically ubiquitous signalling system that regulates diverse aspects of plant development. This system was identified as a direct result of the study of karrikins. Armed with this knowledge, the focus of research in this area will doubtless move towards the wider biological significance and function of ‘the karrikin pathway’. Several key questions remain to be addressed. Most prominently, what is the natural ligand(s) for KAI2, how is it synthesised, and how can we exploit it for beneficial use? The broad-ranging effects of the karrikin pathway provide several possible applications of this knowledge, such as enhancing seed germination rates in recalcitrant species, or boosting seedling establishment under stressful conditions. These applications will require a closer examination of physiological roles of KAI2-dependent signalling, under different growth conditions and in diverse species, so that we can understand precisely what aspects of growth might be manipulated. There are also numerous mysteries pertaining to the molecular mechanisms of the karrikin and strigolactone pathways. What are the specific functions of various SMXL proteins, especially SMAX1 and SMXL2? What are the downstream targets of each SMXL, and how are they distinct from each other? Finally, how does the karrikin pathway facilitate the formation of AM symbiosis? Is it also important in other plant–microbe interactions, such as nodulation or pathogenicity? Understanding these processes in particular will be of major agricultural significance. Answering these enigmas will take time and effort across collaborative and multidisciplinary teams. But without doubt, karrikins have made a permanent imprint upon plant chemical biology.
Acknowledgements
I am grateful to the Australian Society of Plant Scientists for the recognition arising from the award of 2015 Goldacre Medal. I would like to thank several colleagues: Steve Smith, who encouraged me to work with him on karrikins and supported my freedom to develop independent research; Dave Nelson, who initiated the kai screens and continues to be an invaluable collaborator; Christine Beveridge, for mentorship, encouragement and collaboration; Gavin Flematti and Adrian Scaffidi for discussions and essential expertise in all things chemistry; and Yueming Kelly Sun and Nithya Palanivelu, for discussions, innovative ideas and experimental contributions. Finally, I am grateful for funding from the Australian Research Council (grants FT150100162 and DP160102888).
References
Abe S, Sado A, Tanaka K, Kisugi T, Asami K, Ota S, Kim HI, Yoneyama K, Xie X, Ohnishi T, Seto Y, Yamaguchi S, Akiyama K, Yoneyama K, Nomura T (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proceedings of the National Academy of Sciences of the United States of America 111, 18084–18089.| Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhvF2jsL7F&md5=42465fffb2a33b319e91b168e1b44c2dCAS |
Abeles FB, Morgan PW, Saltveit ME, Jr (1992) ‘Ethylene in plant biology.’ (Academic Press: New York)
Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, Greb T (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proceedings of the National Academy of Sciences of the United States of America 108, 20242–20247.
| Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs1yqsb3P&md5=8eb3c4e9be05b50407e65133a02de677CAS |
Akiyama K, Matsuzaki K-I, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827.
| Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXkvVGgsL4%3D&md5=d588b22b5a36c2d71048fbf82fe8b151CAS |
Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annual Review of Plant Biology 66, 161–186.
| Strigolactones, a novel carotenoid-derived plant hormone.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtVajtbvL&md5=696b117dfefa19f8164655f987611e77CAS |
Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164, 711–729.
Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. The Plant Journal 51, 1019–1029.
| DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtFKnur7I&md5=11da8a2f0178ba1bfbc3ad64ffc7a6d0CAS |
Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant & Cell Physiology 50, 1416–1424.
| d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVWitbvP&md5=be0a550989e64902425930c1a8ca419fCAS |
Baldwin IT, Staszak-Kozinski L, Davidson R (1994) Up in smoke: I. Smoke-derived germination cues for postfire annual, Nicotiana attenuata torr. Ex. Watson. Journal of Chemical Ecology 20, 2345–2371.
| Up in smoke: I. Smoke-derived germination cues for postfire annual, Nicotiana attenuata torr. Ex. Watson.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXhtFyhtLo%3D&md5=b7e17a1fa5e2ba95bd7bd7d2d7196107CAS |
Baxter BJM, Baxter BJM, Granger JE, Granger JE, van Staden J, van Staden J (1995) Plant-derived smoke and seed germination: is all smoke good smoke? That is the burning question. South African Journal of Botany 61, 275–277.
| Plant-derived smoke and seed germination: is all smoke good smoke? That is the burning question.Crossref | GoogleScholarGoogle Scholar |
Bennett T, Liang Y, Seale M, Ward S, Müller D, Leyser O (2016) Strigolactone regulates shoot development through a core signalling pathway. Biology Open 5, 1806–1820.
| Strigolactone regulates shoot development through a core signalling pathway.Crossref | GoogleScholarGoogle Scholar |
Beveridge CA, Ross JJ, Murfet IC (1996) Branching in pea (action of genes Rms3 and Rms4). Plant Physiology 110, 859–865.
| Branching in pea (action of genes Rms3 and Rms4).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xhs1GisLY%3D&md5=db30484ff6e376236232c6a684f72d96CAS |
Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C (1997) The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiology 115, 1251–1258.
| The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXns1ykurY%3D&md5=86129b5bfe04757c1b8fce4e612e9c01CAS |
Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology 14, 1232–1238.
| MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmtVams7s%3D&md5=aa37dfcb14db12cb67f8c38f11e9159bCAS |
Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G (2007) Rhizosphere communication of plants, parasitic plants and AM fungi. Trends in Plant Science 12, 224–230.
| Rhizosphere communication of plants, parasitic plants and AM fungi.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXlt1Sitr0%3D&md5=b4fc668226f0a29d86655acca69c655fCAS |
Bravo A, York T, Pumplin N, Mueller LA, Harrison MJ (2016) Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nature Plants 2, 15208
| Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhsVKis7s%3D&md5=af15ba0068d17943f80199539a8b61c3CAS |
Bythell-Douglas R, Waters MT, Scaffidi A, Flematti GR, Smith SM, Bond CS (2013) The structure of the karrikin-insensitive protein (KAI2) in Arabidopsis thaliana. PLoS One 8, e54758
| The structure of the karrikin-insensitive protein (KAI2) in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvFCitbg%3D&md5=996596db1a1a985dfc136464ced6dc00CAS |
Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P (2014) Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. The Plant Cell 26, 1134–1150.
| Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXotV2ktLo%3D&md5=5ef5a0047b6303af4325b475b93b8945CAS |
Conn CE, Nelson DC (2015) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) Receptors may perceive an unknown signal that is not karrikin or strigolactone. Frontiers in Plant Science 6, 1219
| Evidence that KARRIKIN-INSENSITIVE2 (KAI2) Receptors may perceive an unknown signal that is not karrikin or strigolactone.Crossref | GoogleScholarGoogle Scholar |
Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC (2015) Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349, 540–543.
| Convergent evolution of strigolactone perception enabled host detection in parasitic plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXht1egur7O&md5=b3b0d159c88867aad45a742bce43aab9CAS |
Daws MI, Davies J, Pritchard HW, Brown NAC, van Staden J (2007) Butenolide from plant-derived smoke enhances germination and seedling growth of arable weed species. Plant Growth Regulation 51, 73–82.
| Butenolide from plant-derived smoke enhances germination and seedling growth of arable weed species.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXlslWmuw%3D%3D&md5=9dbc06c36878084130c7e49ba8c51585CAS |
de Saint Germain A, Clavé G, Badet-Denisot M-A, Pillot J-P, Cornu D, Le Caer J-P, Burger M, Pelissier F, Retailleau P, Turnbull C, Bonhomme S, Chory J, Rameau C, Boyer FD (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nature Chemical Biology 12, 787–794.
| An histidine covalent receptor and butenolide complex mediates strigolactone perception.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xht1Kmu73J&md5=908ff08748e95c1faeae82b9875069f5CAS |
Delaux P-M, Xie X, Timme RE, Puech-Pagès V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N (2012) Origin of strigolactones in the green lineage. New Phytologist 195, 857–871.
| Origin of strigolactones in the green lineage.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFajtLvN&md5=a296f3a61f8b9315c6707079a3233b71CAS |
Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445.
| The F-box protein TIR1 is an auxin receptor.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXksVeisrs%3D&md5=09ea40f988f4566363ce5c0f83a2b8a0CAS |
Drummond RSM, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC (2009) Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiology 151, 1867–1877.
| Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsFOgtrrK&md5=00805dda959dbb2fca4910cc2a63b077CAS |
Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD (2004) A compound from smoke that promotes seed germination. Science 305, 977
| A compound from smoke that promotes seed germination.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmslWqtrs%3D&md5=8e5514473e8727981589bcf07d7c2bc2CAS |
Flematti GR, Goddard-Borger ED, Merritt DJ, Ghisalberti EL, Dixon KW, Trengove RD (2007) Preparation of 2H-furo[2,3-c]pyran-2-one derivatives and evaluation of their germination-promoting activity. Journal of Agricultural and Food Chemistry 55, 2189–2194.
| Preparation of 2H-furo[2,3-c]pyran-2-one derivatives and evaluation of their germination-promoting activity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhvF2ru70%3D&md5=331fdb130e4a43ceaaa44384badfc555CAS |
Flematti G, Ghisalberti E, Dixon KW, Trengove RD (2008) Germination stimulant in smoke: isolation and identification. ‘Bioactive natural products: detection, isolation, and structural determination’. pp. 531–554. (Taylor & Francis Group: Abingdon, UK)
Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD (2009) Identification of alkyl substituted 2 H-Furo[2,3- c]pyran-2-ones as germination stimulants present in amoke. Journal of Agricultural and Food Chemistry 57, 9475–9480.
| Identification of alkyl substituted 2 H-Furo[2,3- c]pyran-2-ones as germination stimulants present in amoke.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtFOqtb%2FI&md5=737f714d46a15e46ae12979bfb1d73b3CAS |
Flematti GR, Merritt DJ, Piggott MJ, Trengove RD, Smith SM, Dixon KW, Ghisalberti EL (2011) Burning vegetation produces cyanohydrins that liberate cyanide and stimulate seed germination. Nature Communications 2, 360
| Burning vegetation produces cyanohydrins that liberate cyanide and stimulate seed germination.Crossref | GoogleScholarGoogle Scholar |
Flematti GR, Scaffidi A, Waters MT, Smith SM (2016) Stereospecificity in strigolactone biosynthesis and perception. Planta 243, 1361–1373.
| Stereospecificity in strigolactone biosynthesis and perception.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xms1OjsL8%3D&md5=694e3a8d7048d372f46a8c436fe525fdCAS |
Foo E, Yoneyama K, Hugill CJ, Quittenden LJ, Reid JB (2013) Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Molecular Plant 6, 76–87.
| Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtFCntbg%3D&md5=42420265724cee406cc1e5ef60a2579fCAS |
Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C, Bouwmeester H, Bécard G, Beveridge CA, Rameau C, Rochange SF (2008) Strigolactone inhibition of shoot branching. Nature 455, 189–194.
| Strigolactone inhibition of shoot branching.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtV2qtLzK&md5=648081257304ee7344e5535e41538f6aCAS |
Guo Y, Zheng Z, La Clair JJ, Chory J, Noel JP (2013) Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 110, 8284–8289.
| Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtV2ntLbO&md5=abb030a6101f92e79fc18216317d8907CAS |
Gutjahr C, Gobbato E, Choi J, Riemann M, Johnston MG, Summers W, Carbonnel S, Mansfield C, Yang S-Y, Nadal M, Acosta I, Takano M, Jiao WB, Schneeberger K, Kelly KA, Paszkowski U (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350, 1521–1524.
| Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXitVWrtLzF&md5=ffaa1b3bd61b666436b5c10959357619CAS |
Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology 22, 2032–2036.
| DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtlSktL7L&md5=2f00ae2815824855956a9008d3afd5bdCAS |
Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant & Cell Physiology 46, 79–86.
| Suppression of tiller bud activity in tillering dwarf mutants of rice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXht1eqt7o%3D&md5=7e0acd496d9c8df1b728548b167ef2b9CAS |
Jain N, Kulkarni MG, Van Staden J (2006) A butenolide, isolated from smoke, can overcome the detrimental effects of extreme temperatures during tomato seed germination. Plant Growth Regulation 49, 263–267.
| A butenolide, isolated from smoke, can overcome the detrimental effects of extreme temperatures during tomato seed germination.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtlalsL3M&md5=622ce3365b76b4b3f8dac4aa93739371CAS |
Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, Yi W, Zhao L, Ma H, He Y, Wu Z, Melcher K, Qian Q, Xu HE, Wang Y, Li J (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405.
| DWARF 53 acts as a repressor of strigolactone signalling in rice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvFOmtrrK&md5=fd87712347d5a0c98cf34c71ff910f69CAS |
Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C (2006) Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiology 142, 1014–1026.
| Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1ejurfN&md5=3e4f050f838ff909e906eacf2f1d3de9CAS |
Kagiyama M, Hirano Y, Mori T, Kim S-Y, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes to Cells 18, 147–160.
| Structures of D14 and D14L in the strigolactone and karrikin signaling pathways.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtlajurY%3D&md5=6b6d9076c12c6e4cb77f02095005ae2cCAS |
Kameoka H, Kyozuka J (2015) Downregulation of rice DWARF 14 LIKE suppress mesocotyl elongation via a strigolactone independent pathway in the dark. Journal of Genetics and Genomics 42, 119–124.
| Downregulation of rice DWARF 14 LIKE suppress mesocotyl elongation via a strigolactone independent pathway in the dark.Crossref | GoogleScholarGoogle Scholar |
Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proceedings of the National Academy of Sciences of the United States of America 105, 7100–7105.
| COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXmt1Gmurc%3D&md5=d7bd7fcf7c18305fba2a5d2de31d21cbCAS |
Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451.
| The Arabidopsis F-box protein TIR1 is an auxin receptor.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXksVeisro%3D&md5=fd2e188c7b17bd3134b032a2969a86b0CAS |
Khosla A, Nelson DC (2016) Strigolactones, super hormones in the fight against Striga. Current Opinion in Plant Biology 33, 57–63.
| Strigolactones, super hormones in the fight against Striga.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XpvVWku7g%3D&md5=78f83f0e3dfca221c4cbed3739a76cd8CAS |
Kochanek J, Long RL, Lisle AT, Flematti GR (2016) Karrikins identified in biochars indicate post-fire chemical cues can influence community diversity and plant development. PLoS One 11, e0161234
| Karrikins identified in biochars indicate post-fire chemical cues can influence community diversity and plant development.Crossref | GoogleScholarGoogle Scholar |
Kohlen W, Charnikhova T, Lammers M, Pollina T, Tóth P, Haider I, Pozo MJ, de Maagd RA, Ruyter-Spira C, Bouwmeester HJ, López-Ráez JA (2012) The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytologist
| The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis.Crossref | GoogleScholarGoogle Scholar |
Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483, 341–344.
| A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjtlOgurc%3D&md5=225bb21bba86e2322007a325234621e6CAS |
Kulkarni MG, Sparg SG, Light ME (2006) Stimulation of rice (Oryza sativa L.) seedling vigour by smoke‐water and butenolide. Journal of Agronomy 192, 395–398.
| Stimulation of rice (Oryza sativa L.) seedling vigour by smoke‐water and butenolide.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFOrsbzP&md5=1e46c6018be2d045a0cc76c744dac862CAS |
Liang Y, Ward S, Li P, Bennett T, Leyser O (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. The Plant Cell 28, 1581–1601.
| SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhslCqurnP&md5=b9d84a62f9afb50a392ba5b9a13f8626CAS |
Liu W, Wu C, Fu Y, Hu G, Si H, Zhu L, Luan W, He Z, Sun Z (2009) Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230, 649–658.
| Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVait7zF&md5=9efbfdaf45ba71556026ce4ca82bb5edCAS |
Long RL, Williams K, Griffiths EM, Flematti GR, Merritt DJ, Stevens JC, Turner SR, Powles SB, Dixon KW (2010) Prior hydration of Brassica tournefortii seeds reduces the stimulatory effect of karrikinolide on germination and increases seed sensitivity to abscisic acid. Annals of Botany 105, 1063–1070.
| Prior hydration of Brassica tournefortii seeds reduces the stimulatory effect of karrikinolide on germination and increases seed sensitivity to abscisic acid.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXmvVWltLc%3D&md5=09645ac753d7a0ec9e7119a6b516e37fCAS |
Long RL, Stevens JC, Griffiths EM, Adamek M, Gorecki MJ, Powles SB, Merritt DJ (2011) Seeds of Brassicaceae weeds have an inherent or inducible response to the germination stimulant karrikinolide. Annals of Botany 108, 933–944.
| Seeds of Brassicaceae weeds have an inherent or inducible response to the germination stimulant karrikinolide.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1eisbrL&md5=144d8fd2ca284d0fe580862883d63e9bCAS |
Lopez-Obando M, Conn CE, Hoffmann B, Bythell-Douglas R, Nelson DC, Rameau C, Bonhomme S (2016) Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens. Planta 243, 1441–1453.
| Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XktlChtL4%3D&md5=1218daa5ec3239c3a84ee81e001c2badCAS |
Mander LN (2003) Twenty years of gibberellin research. Natural Product Reports 20, 49–69.
| Twenty years of gibberellin research.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXisVKls70%3D&md5=245c76e33f5e113c32c6a6ed20dd1973CAS |
Matusova R, Rani K, Verstappen F, Franssen M, Beale M, Bouwmeester H (2005) The strigolactone germination stimulants of the plant–parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiology 139, 920
| The strigolactone germination stimulants of the plant–parasitic Striga and Orobanche spp. are derived from the carotenoid pathway.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtFCgsb%2FI&md5=08c70895c57e5d8ef40e5f40af304559CAS |
Mayzlish-Gati E, De Cuyper C, Goormachtig S, Beeckman T, Vuylsteke M, Brewer PB, Beveridge CA, Yermiyahu U, Kaplan Y, Enzer Y, Wininger S, Resnick N, Cohen M, Kapulnik Y, Koltai H (2012) Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiology 160, 1329–1341.
| Strigolactones are involved in root response to low phosphate conditions in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs12kurvO&md5=3f22fba3e9464fedbaba178425b70234CAS |
McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-P, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. The Plant Cell 15, 1120–1130.
| The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjvV2htLY%3D&md5=4e50950476b2ad4ded1158fcf5fc329fCAS |
Nakamura H, Xue Y-L, Miyakawa T, Hou F, Qin H-M, Fukui K, Shi X, Ito E, Ito S, Park S-H, Miyauchi Y, Asano A, Totsuka N, Ueda T, Tanokura M, Asami T (2013) Molecular mechanism of strigolactone perception by DWARF14. Nature Communications 4, 2613
| Molecular mechanism of strigolactone perception by DWARF14.Crossref | GoogleScholarGoogle Scholar |
Napoli C (1996) Highly branched phenotype of the Petunia dad1-1 mutant is reversed by grafting. Plant Physiology 111, 27–37.
| Highly branched phenotype of the Petunia dad1-1 mutant is reversed by grafting.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XivFektLw%3D&md5=d4bd36e68d3f6238ea7942bcdb246412CAS |
Nelson DC, Riseborough J-A, Flematti GR, Stevens J, Ghisalberti EL, Dixon KW, Smith SM (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiology 149, 863–873.
| Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXjt1Wqu7s%3D&md5=877dd3742b604f1aa387ce88c9328391CAS |
Nelson DC, Flematti GR, Riseborough J-A, Ghisalberti EL, Dixon KW, Smith SM (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 107, 7095–7100.
| Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXltFSjtbo%3D&md5=4a3776a7a100d5d2077386faf4be8b6fCAS |
Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 108, 8897–8902.
| F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXntVektLg%3D&md5=a35324635e5f7bb9cc1c9c74f806826eCAS |
Nelson DC, Flematti GR, Ghisalberti EL, Dixon KW, Smith SM (2012) Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annual Review of Plant Biology 63, 107–130.
| Regulation of seed germination and seedling growth by chemical signals from burning vegetation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xos1amsrc%3D&md5=ceac557740bf1a886f87e9d4fb939d0cCAS |
Parker C (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Management Science 65, 453–459.
| Observations on the current status of Orobanche and Striga problems worldwide.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXlsFymurw%3D&md5=332c439ddf6cb3310571b38526a959a1CAS |
Pausas JG, Keeley JE (2009) A burning story: the role of fire in the history of life. Bioscience 59, 593–601.
| A burning story: the role of fire in the history of life.Crossref | GoogleScholarGoogle Scholar |
Rasmussen A, Beveridge CA, Geelen D (2012) Inhibition of strigolactones promotes adventitious root formation. Plant Signaling & Behavior 7, 694–697.
| Inhibition of strigolactones promotes adventitious root formation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhslSktrzI&md5=ebb772e71cd1cbf67df1fa40d8acad9cCAS |
Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, Verstappen F, Bouwmeester H (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiology 155, 721–734.
| Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXksFOjsLk%3D&md5=3971d3f129bc1ad3775d0247ab8760fcCAS |
Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H (2013) The biology of strigolactones. Trends in Plant Science 18, 72–83.
| The biology of strigolactones.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhslSmtLjK&md5=ea27aff18dc569c3eb9ef105c311eeecCAS |
Scaffidi A, Waters MT, Bond CS, Dixon KW, Smith SM, Ghisalberti EL, Flematti GR (2012) Exploring the molecular mechanism of karrikins and strigolactones. Bioorganic & Medicinal Chemistry 22, 3743–3746.
| Exploring the molecular mechanism of karrikins and strigolactones.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XmtlWlsrk%3D&md5=52bc29447aa0c4a1e405b986cd79998fCAS |
Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiology 165, 1221–1232.
| Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtFOqsbvE&md5=596b31d47058dd5f0f4d91778aded561CAS |
Scott AC, Glasspool IJ (2006) The diversification of Paleozoic fire systems and fluctuations in atmospheric oxygen concentration. Proceedings of the National Academy of Sciences of the United States of America 103, 10861–10865.
| The diversification of Paleozoic fire systems and fluctuations in atmospheric oxygen concentration.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XnsFegu7g%3D&md5=ad0e5f38fbe05b1f067ad62010cfcef6CAS |
Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ (2005) The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. The Plant Cell 17, 746–759.
| The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXis1ygtr0%3D&md5=938291f842afce3fa07ccb8ee0d20d9fCAS |
Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, Leyser O (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes & Development 17, 1469–1474.
| MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXkvFSisrk%3D&md5=9f4d6d4647f40f267f53979467c7d699CAS |
Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. The Plant Cell 27, 3143–3159.
| SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhsVantrrO&md5=5a82b76f8425f518f561964dc6dcdff7CAS |
Stanga JP, Smith SM, Briggs WR, Nelson DC (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiology 163, 318–330.
| SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsVygsbnO&md5=a59e33569ece7d38dec1ed3c65689d1eCAS |
Stanga JP, Morffy N, Nelson DC (2016) Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243, 1397–1406.
| Functional redundancy in the control of seedling growth by the karrikin signaling pathway.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xmslymuw%3D%3D&md5=55501e99108cff689abbc61b20d97a3aCAS |
Stevens J, Merritt D, Flematti G, Ghisalberti E, Dixon K (2007) Seed germination of agricultural weeds is promoted by the butenolide 3-methyl-2H-furo[2,3-c]pyran-2-one under laboratory and field conditions. Plant and Soil 298, 113–124.
| Seed germination of agricultural weeds is promoted by the butenolide 3-methyl-2H-furo[2,3-c]pyran-2-one under laboratory and field conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtVejtr3L&md5=2d15341f643fe65443bc79b4bdd23d3fCAS |
Stirnberg P, van De Sande K, Leyser HMO (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141.
Stirnberg P, Zhao S, Williamson L, Ward S, Leyser O (2012) FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. The Plant Journal 71, 907–920.
| FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtlSmt77J&md5=5fc1ab034905bbcf7e0298e8b24451d6CAS |
Sun X-D, Ni M (2011) HYPOSENSITIVE TO LIGHT, an α/β fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Molecular Plant 4, 116–126.
| HYPOSENSITIVE TO LIGHT, an α/β fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlemu74%3D&md5=388556ec36e520fe079a8d1faf318c03CAS |
Sun YK, Flematti GR, Smith SM, Waters MT (2016) Reporter gene-facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway. Frontiers in Plant Science 7, 1799
| Reporter gene-facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway.Crossref | GoogleScholarGoogle Scholar |
Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207.
| Structure-function analysis identifies highly sensitive strigolactone receptors in Striga.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhs1antLnI&md5=13c533338278db3cae5802e6f51c1fc3CAS |
Tsuchiya Y, Yoshimura M, Sato Y, Kuwata K, Toh S, Holbrook-Smith D, Zhang H, McCourt P, Itami K, Kinoshita T, Hagihara S (2015) Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868.
| Probing strigolactone receptors in Striga hermonthica with fluorescence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtlKlt7%2FP&md5=d6c75ac46ca44339e825db0c4eeeb062CAS |
Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow T-Y, Hsing Y-IC, Kitano H, Yamaguchi I, Matsuoka M (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698.
| GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVCjsL%2FP&md5=7ef1ed8c628cd797211e0aa5d67fce2eCAS |
Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200.
| Inhibition of shoot branching by new terpenoid plant hormones.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtV2qtLnE&md5=aa33e00ac2d0ef479b6f245c18394ddaCAS |
van der Schaar W, Alonso-Blanco C, Léon-Kloosterziel KM, Jansen RC, van Ooijen JW, Koornneef M (1997) QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity 79, 190–200.
| QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping.Crossref | GoogleScholarGoogle Scholar |
van Staden J, van Staden J, Drewes FE, Drewes FE, Brown NAC, Brown NAC (1995) Some chromatographic characteristics of germination stimulants in plant-derived smoke extracts. Plant Growth Regulation 17, 241–249.
| Some chromatographic characteristics of germination stimulants in plant-derived smoke extracts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXpvVOjtr0%3D&md5=6fe12cbb3d7eda66394bbe7dd4fa05b1CAS |
Van Staden J, Brown NAC, Jger AK, Johnson TA (2000) Smoke as a germination cue. Plant Species Biology 15, 167–178.
| Smoke as a germination cue.Crossref | GoogleScholarGoogle Scholar |
van Staden J, Jager AK, Light ME, Burger BV (2004) Isolation of the major germination cue from plant-derived smoke. South African Journal of Botany 70, 654–659.
| Isolation of the major germination cue from plant-derived smoke.Crossref | GoogleScholarGoogle Scholar |
van Staden J, Sparg S, Kulkarni M, Light M (2006) Post-germination effects of the smoke-derived compound 3-methyl-2H-furo [2, 3-c] pyran-2-one, and its potential as a preconditioning agent. Field Crops Research 98, 98–105.
| Post-germination effects of the smoke-derived compound 3-methyl-2H-furo [2, 3-c] pyran-2-one, and its potential as a preconditioning agent.Crossref | GoogleScholarGoogle Scholar |
Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. The Plant Cell 27, 3128–3142.
| Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XnsFKntbg%3D&md5=125e026c091c14f3f3de49d193f6786aCAS |
Waters MT, Smith SM (2013) KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Molecular Plant 6, 63–75.
| KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtFCnurc%3D&md5=8397b17016cfca77a4e1727de33146beCAS |
Waters MT, Smith SM, Nelson DC (2011) Smoke signals and seed dormancy: where next for MAX2? Plant Signaling & Behavior 6, 1418–1422.
| Smoke signals and seed dormancy: where next for MAX2?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjtlOgsL8%3D&md5=ad4665d2f162bd1c4427f07ee24690b3CAS |
Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012a) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295.
| Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XmsFyqt7k%3D&md5=5ffec61ce46de74d54f93195b393ed3fCAS |
Waters MT, Scaffidi A, Flematti GR, Smith SM (2012b) Karrikins force a rethink of strigolactone mode of action. Plant Signaling & Behavior 7, 969–972.
| Karrikins force a rethink of strigolactone mode of action.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtFGgtLs%3D&md5=7b19e9f8dfec477293b744307a936d74CAS |
Waters MT, Scaffidi A, Flematti GR, Smith SM (2013) The origins and mechanisms of karrikin signalling. Current Opinion in Plant Biology 16, 667–673.
| The origins and mechanisms of karrikin signalling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXht12isrbN&md5=885800159b14127d3882a9b2e2598525CAS |
Waters MT, Scaffidi A, Sun YK, Flematti GR, Smith SM (2014) The karrikin response system of Arabidopsis. The Plant Journal 79, 623–631.
| The karrikin response system of Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtlaktb3O&md5=030b387c60ecc08e1d8497d46abc5232CAS |
Waters MT, Scaffidi A, Flematti G, Smith SM (2015a) Substrate-induced degradation of the α/β-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Molecular Plant 8, 814–817.
| Substrate-induced degradation of the α/β-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXpsFeks7o%3D&md5=045723798f427375c6875fbe0d5ce54bCAS |
Waters MT, Scaffidi A, Moulin SLY, Sun YK, Flematti GR, Smith SM (2015b) A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. The Plant Cell 27, 1925–1944.
| A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhsVOhsbfF&md5=d39c47b98eb48fe802b6a61835eefd67CAS |
Waters MT, Gutjahr C, Bennett T, Nelson DC (2017) Strigolactone signaling and evolution. Annual Review of Plant Biology
| Strigolactone signaling and evolution.Crossref | GoogleScholarGoogle Scholar |
Xu G, Ma H, Nei M, Kong H (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proceedings of the National Academy of Sciences of the United States of America 106, 835–840.
| Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXht12isbg%3D&md5=403f55eb8fa7de364c5e3522e24d75d0CAS |
Xu Y, Miyakawa T, Nakamura H, Nakamura A, Imamura Y, Asami T, Tanokura M (2016) Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica. Scientific Reports 6, 31386
| Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhtlCitrnP&md5=9942d3dfc25c7c5c48aea6f85c845bb3CAS |
Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta
| Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency.Crossref | GoogleScholarGoogle Scholar |
Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, et al (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473.
| DWARF14 is a non-canonical hormone receptor for strigolactone.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xht1GrurfM&md5=13f1d37ef182e3c89bc36950c7e72e1fCAS |
Yoshida S, Kameoka H, Tempo M, Akiyama K, Umehara M, Yamaguchi S, Hayashi H, Kyozuka J, Shirasu K (2012) The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytologist 196, 1208–1216.
| The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1WjurzP&md5=da301a5d6697ed855fbf5a7df1d10ca0CAS |
Zhao L-H, Zhou XE, Wu Z-S, Yi W, Xu Y, Li S, Xu T-H, Liu Y, Chen R-Z, Kovach A, Kang Y, et al (2013) Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Research 23, 436–439.
| Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjsFaqtbg%3D&md5=09ad0f09a789851128c3c67302c6abd3CAS |
Zhao L-H, Zhou XE, Yi W, Wu Z, Liu Y, Kang Y, Hou L, de Waal PW, Li S, Jiang Y, et al (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Research 25, 1219–1236.
| Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhslWlurfJ&md5=0d6408aee1bc3728046af7c29209e572CAS |
Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al (2013) D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410.
| D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvFOls7nL&md5=be03aa9868f178e8663664de3d36bfadCAS |
Zhou J, da Silva J, Ma G (2014) Effects of smoke water and karrikin on seed germination of 13 species growing in China. Central European Journal of Biology 9, 1108–1116.
| Effects of smoke water and karrikin on seed germination of 13 species growing in China.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsValtLbE&md5=a7eadd6d4a4740922a7b03a3ce87aec9CAS |


