264 ANALYSIS OF DNA METHYLATION PATTERN IN PRE-IMPLANTATION-STAGE EMBRYOS DERIVED FROM NUCLEAR TRANSFER USING PORCINE EMBRYONIC GERM CELLS
D.-H. Choi A , C.-H. Park A , S.-G. Lee A , H.-S. Kim A , H.-Y. Son A and C.-K. Lee AACollege of Agriculture and Life Science, Seoul National University, Seoul, Korea
Reproduction, Fertility and Development 19(1) 248-249 https://doi.org/10.1071/RDv19n1Ab264
Submitted: 12 October 2006 Accepted: 12 October 2006 Published: 12 December 2006
Abstract
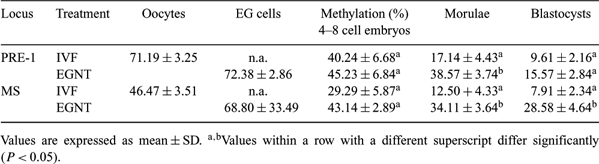
Somatic cell nuclear transfer (SCNT) has been successfully used to produce live cloned offspring in various mammals. However, some studies had reported that cloned embryos by SCNT had many problems in reprogramming or epigenetic modification, such as DNA methylation. DNA methylation is an essential process in epigenetic modification for development, and aberrant methylation in cloned embryos gives rise to abortion, high birth weight, and perinatal death. In this study, embryonic germ (EG) cells were used as donor cells for nuclear transfer. EG cells may have less reprogramming or demethylation than SCNT because these are already in erased status. However, little is known about methylation state or developmental capacity of the EG cell as a donor. The objective of this study was to analyze the methylation pattern of pre-implantation embryos cloned from porcine EG cells. Two regions, PRE-1 and microsatellite (MS), were analyzed for methylation patterns of cloned embryos from porcine EG cells and compared with the pattern of mature oocytes and in vitro-fertilized (IVF) embryos as a control. Cumulus–oocyte complexes were collected from prepubertal gilt ovaries and matured in vitro for 44 h, followed by use for IVF and NT with porcine EG cells. The porcine EG cells were prepared from 28-day-old fetuses after mating; genital ridges were isolated from fetuses, and then transferred into a culture medium on a feeder layer. The number of embryos for analysis was 300 for matured oocytes, 50–80 for 4–8 cell embryos, 30–40 for morulae, and 20–30 for blastocysts. The genomic DNA was isolated from the embryos and treated with bisulfite solution. PCR was performed for the amplification of PRE-1 and MS regions. The PCR products were sequenced by using an automatic DNA sequencer. The methylation rates of the PRE-1 and MS regions in IVF embryos showed that the demethylation process had occurred during the pre-implantation stage, which is a typical phenomenon of in vivo counterparts (Kang et al. 2001 J. Biol. Chem. 276, 39 980). However, compared to IVF embryos, embryos derived from NT using EG cells showed differences at the morula (PRE-1) and blastocyst (MS) stage. These results indicate that porcine EG cells also have problems in reprogramming during NT. For detailed and reliable results, the methylation pattern analysis of the imprinting region, for example, H19 in maternal allele and Igf2 in paternal allele, must be examined.
|


