Molecular control of mitochondrial function in developing rhesus monkey oocytes and preimplantation-stage embryos
N. R. Mtango A E , A. J. Harvey C E , K. E. Latham A B and C. A. Brenner C D FA The Fels Institute for Cancer Research and Molecular Biology, Temple University School of Medicine, Philadelphia, PA 19140, USA.
B Department of Biochemistry, Temple University School of Medicine, Philadelphia, PA 19140, USA.
C Department of Physiology, Wayne State University, School of Medicine, Detroit, MI 48201, USA.
D Department of Obstetrics and Gynecology, Wayne State University, School of Medicine, Detroit, MI 48201, USA.
E These authors contributed equally to this study.
F Corresponding author. Email: cbrenner@med.wayne.edu
Reproduction, Fertility and Development 20(7) 846-859 https://doi.org/10.1071/RD08078
Submitted: 18 April 2008 Accepted: 21 July 2008 Published: 16 September 2008
Abstract
The mitochondrion undergoes significant functional and structural changes, as well as an increase in number, during preimplantation embryonic development. The mitochondrion generates ATP and regulates a range of cellular processes, such as signal transduction and apoptosis. Therefore, mitochondria contribute to overall oocyte quality and embryo developmental competence. The present study identified, for the first time, the detailed temporal expression of mRNAs related to mitochondrial biogenesis in rhesus monkey oocytes and embryos. Persistent expression of maternally encoded mRNAs was observed, in combination with transcriptional activation and mRNA accumulation at the eight-cell stage, around the time of embryonic genome activation. The expression of these transcripts was significantly altered in oocytes and embryos with reduced developmental potential. In these embryos, most maternally encoded transcripts were precociously depleted. Embryo culture and specific culture media affected the expression of some of these transcripts, including a deficiency in the expression of key transcriptional regulators. Several genes involved in regulating mitochondrial transcription and replication are similarly affected by in vitro conditions and their downregulation may be instrumental in maintaining the mRNA profiles of mitochondrially encoded genes observed in the present study. These data support the hypothesis that the molecular control of mitochondrial biogenesis, and therefore mitochondrial function, is impaired in in vitro-cultured embryos. These results highlight the need for additional studies in human and non-human primate model species to determine how mitochondrial biogenesis can be altered by oocyte and embryo manipulation protocols and whether this affects physiological function in progeny.
Additional keywords: gene regulation, macaque, mitochondrial DNA transcription factors.
Introduction
Preimplantation embryos are dependent on the energy produced from oocyte-inherited mitochondria. The mitochondrion is a key player in several cellular processes in addition to ATP production through oxidative phosphorylation (OXPHOS), including ion homeostasis, the synthesis of metabolites and the modulation of signal transduction. These mitochondrial functions are critical determinants of early embryonic development at various levels, including spindle organisation, chromosomal segregation and cell cycle regulation, as well as during processes such as compaction, cavitation and blastocyst hatching (Wilding et al. 2001; Van Blerkom 2004; Thundathil et al. 2005).
The distribution of mitochondria within a cell varies with the stage of the cell cycle (Margineantu et al. 2002) and may play an important regulatory role in nuclear–mitochondrial signalling. The appropriate distribution of mitochondria contributes to organelle function and is essential for cell survival (Frederick and Shaw 2007). Mitochondrial localisation, structure and activity change significantly during mammalian oogenesis and early embryogenesis (Bavister and Squirrell 2000), with several studies indicating that mitochondrial organisation is associated with developmental competence (Van Blerkom et al. 1990; Barnett et al. 1997; Lane and Bavister 1998; Ludwig et al. 2001; Squirrell et al. 2001, 2003). Active mitochondria relocate during oocyte maturation and fertilisation in most species. Both rhesus and human zygotes display a distinct accumulation of mitochondria between the pronuclei before syngamy (Van Blerkom et al. 2000; Squirrell et al. 2003). Mitochondrial morphology also changes significantly during the period of preimplantation development. Metaphase (M) II oocytes contain small spherical mitochondria with few undeveloped cristae that surround electron-dense matrices (Van Blerkom 2004). In the mouse, during development from the four- to eight-cell stage through to the blastocyst stage, these mitochondria become elongated with numerous and predominantly transversely arranged cristae (Piko and Matsumoto 1976). Changes in mitochondrial structure are associated with increased mitochondrial gene expression (Piko and Taylor 1987), the activation and transcription of mitochondrial and nuclear genes (Taylor and Piko 1995), increased oxygen consumption and ATP production via OXPHOS (Trimarchi et al. 2000; Sturmey and Leese 2003; Van Blerkom 2004) and an increase in the abundance of mitochondria with high membrane potential (Acton et al. 2004).
Each mitochondrion contains several copies of mitochondrial (mt) DNA. The mtDNA content varies considerably in human or primate oocytes, even when oocytes are collected from the same individual. The number of mtDNA copies in MII oocytes has been reported to differ by well over an order of magnitude, ranging from 2 × 104 to over 8 × 105 (Steuerwald et al. 2000a, 2000b; Reynier et al. 2001; Barritt et al. 2002). Mitochondrial function and biogenesis rely on the intricate coordination and expression of both nuclear and mitochondrial genomes (Garesse and Vallejo 2001; Harvey et al. 2007; Scarpulla 2008). The mtDNA spans an approximate 16.5 kb circular DNA molecule, encoding 13 proteins involved in the electron transport chain, two rRNAs and 22 tRNAs. The heavy strand encodes for 28 genes, whereas the light strand encodes for nine genes, eight of which are tRNAs (Fig. 1). The mtDNA lacks introns, with an approximate 1200-nucleotide non-coding region (D-loop; displacement loop or control region), which contains the origins of replication for each strand (OH and OL for the heavy and light strands, respectively). Although the mechanisms that regulate mitochondrial function in somatic cells have started to be elucidated (Shadel and Clayton 1997; Moraes 2001), the regulation of mitochondrial function during preimplantation embryo development remains largely unknown.
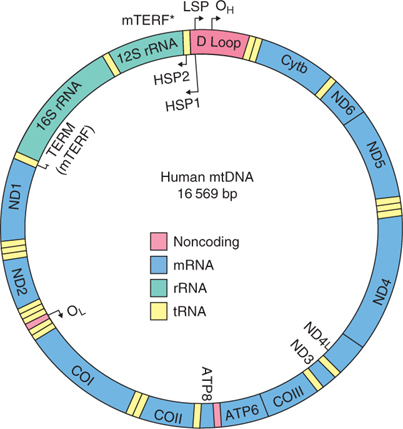
|
Both the heavy and light strands of mtDNA contain a single promoter region for transcriptional initiation. Transcription of the heavy strand is initiated from two specific sites, namely HSP1 and HSP2, whereas LSP represents the initiation site for the light strand (for a review, see Scarpulla 2008). HSP1 transcribes the entire heavy strand, producing a polycistronic transcript that corresponds to the full-length mtDNA molecule, whereas HSP2 results in the transcription of the two mitochondrial rRNA molecules (Asin-Cayuela and Gustafsson 2007). Transcription from LSP transcribes the entire light strand, but also produces the RNA primers required for initiation of mtDNA replication at the origin of H-strand DNA replication (OH; Falkenberg et al. 2007). The process of transcription involves mitochondrial RNA polymerase (POLRMT), mitochondrial transcription factor A (TFAM) and mitochondrial transcription factors B1 and B2 (TFB1M and TFB2M, respectively). A target sequence immediately upstream from the transcription promoter serves as the binding site for nuclear-encoded TFAM. TFAM, a member of the high-mobility group of proteins, is imported to mitochondria, where it functions as a key regulator of mammalian mitochondrial transcription by bending and unwinding mtDNA upon binding to its target sequence in the promoter region (Chang and Clayton 1984). Transcription initiation requires POLRMT, which is responsible for site-specific promoter recognition (Falkenberg et al. 2007) and uses TFB1M or TFB2M for promoter selection and transcriptional specificity (Falkenberg et al. 2002; McCulloch et al. 2002).
Replicaton of mtDNA occurs as an asynchronous displacement process involving two independent origins. Replication is initiated on the heavy strand at the origin of replication (OH) in the D-loop, which is downstream from the LSP, thereby displacing the other strand. When H-strand replication exposes the light strand origin of replication (OL), replication is initiated and proceeds in the opposite direction (Lee and Clayton 1996; Shadel and Clayton 1997). Replicaton of mtDNA is initiated by mitochondria-specific DNA polymerase gamma (POLG). POLG is composed of two subunits, a large catalytic α-subunit (Gray and Wong 1992; Graves et al. 1998) that also has exonuclease activity (Longley et al. 2001), and a smaller accessory subunit (POLGB) that appears to be involved in primer recognition and confers improved polymerase capabilities in combination with POLGA (Carrodeguas et al. 1999; Fan et al. 1999; Lim et al. 1999). Mitochondrial single-strand binding protein (SSBP1) maintains the integrity and increases activity and fidelity of POLG (Hoke et al. 1990; Farr et al. 1999). TFAM, in addition to being required for in vitro transcription initiation, appears to have a direct role in mtDNA maintenance (Evans and Scarpulla 1990). Other key players include nuclear respiratory factor (NRF) 1 and NRF2, which regulate mtDNA transcription (Virbasius and Scarpulla 1994). It is assumed that mtDNA replication does not occur after oocyte growth until the blastocyst stage (Piko and Taylor 1987; Smith and Alcivar 1993).
Previous temporal studies of mouse and bovine embryo development revealed that nuclear-encoded mitochondrial transcription factors are associated with an upregulation of mitochondrial transcription in morula and blastocyst-stage embryos (May-Panloup et al. 2005; Thundathil et al. 2005; Bowles et al. 2007; Spikings et al. 2007). Although transcripts are abundant during this time, mtDNA replication does not occur until the blastocyst stage in the mouse (Thundathil et al. 2005). However, understanding of the molecular events controlling mitochondrial function during primate embryo development remains rudimentary. Therefore, to determine mRNA steady state levels of mitochondrial genes (mtDNA transcription) and nuclear genes encoding factors involved in mitochondrial transcription and replication, a quantitative analysis of mRNAs was undertaken in the present study.
Materials and methods
Oocytes and embryos
The isolation and culture of rhesus oocytes and embryos was performed during construction of the Primate Embryo Gene Expression Resource (PREGER; http://www.preger.org), as described previously (Zheng et al. 2004). PREGER contains more than 170 samples of rhesus monkey oocytes and preimplantation-stage embryos. The oocytes used to determine the effect of hormonal stimulation on gene expression were obtained from monkeys that were unstimulated (denoted as ‘NS’) or stimulated with either follicle-stimulating hormone (FSH) only or FSH followed by human chorionic gonadotropin (FSH+hCG) and matured either in vitro (NS and FSH) or in vivo (FSH+hCG). In addition, pronuclear and two-cell embryos derived from these three kinds of oocytes by in vitro fertilisation (IVF) and cultured in HECM9 medium (Schramm and Bavister 1995, 1996; McKiernan and Bavister 2000) were analysed. Additional data from embryos (greater than two-cell stage) developing from NS and FSH-stimulated oocytes are available at http://www.preger.org.
Temporal gene expression patterns were determined from oocytes and embryos cultured under standard IVF/HECM9 conditions from FSH+hCG-stimulated females, with the exception of germinal vesicle (GV)-stage oocytes, which were collected from females stimulated with FSH only. Embryos treated with 24 μg mL–1 of the transcriptional inhibitor α-amanitin were included in the sample set to determine whether RNA synthesis of any transcript was inhibited around the time of embryonic genome activation. Samples of embryos from normal cycles and flushed from the reproductive tract without culture were collected to determine the effect of culture on gene expression patterns. All somatic cells and the zona pellucida were removed from oocytes and embryos before processing for reverse transcription–polymerase chain reaction (RT-PCR). For all stages and conditions, between three and thirteen samples were obtained from one to four oocytes or embryos, with the exception of G1/G2 cultured blastocysts, for which only two samples were available for analysis. All oocytes and embryos collected for inclusion in the PREGER sample database were of high quality and healthy in appearance.
Samples were collected in accordance with The Wisconsin National Primate Research Center Animal Care and Use Committee at the time of PREGER construction. The animals were maintained according to recommendations of the Guide for the Care and Use of Laboratory Animals.
Analysis of mRNA expression
Primers were designed using Primer Express (Applied Biosystems, Foster City, CA, USA) or PRIMER3 Software (Rozen and Skaletsky 2000; Table 1). Complementary DNA probes were obtained by PCR from specific clones obtained from Open Biosystems (Huntsville, AL, USA) with the exception of MT-CYB, MT-CO1, MT-CO2, MT-ND2 and MTIF2 (Table 2), which were generated by RT-PCR using Rhesus Monkey Skeletal Muscle RNA (BioChain, Hayward, CA, USA). Identities of the amplified cDNAs were confirmed by DNA sequencing. Blot preparation, probe preparation, hybridisation and quantitative analyses were performed as described previously (Rambhatla et al. 1995; Latham et al. 1999; Zheng et al. 2004).

|

|
Data are expressed as the mean ± s.e.m. counts per minute (c.p.m.) bound for each culture condition or the stage of oocyte or embryo development. Statistical differences in hybridisation signals obtained for different stages or conditions were evaluated by Student’s t-test. Corrections for multiple testing were incorporated into the comparisons between oocytes obtained from NS and FSH- and FSH+hCG-stimulated females and embryos derived from these oocytes. To provide a complete overview of the effects of these different protocols, annotations for both the corrected (P < 0.016) and uncorrected (P < 0.05) confidence levels are provided.
Results
The PREGER resource was used to examine the expression of mitochondrial- and nuclear-encoded genes involved in the regulation of mtDNA replication and transcription. The PREGER resource contains samples of oocytes and embryos obtained using diverse protocols, encompassing in vivo or in vitro maturation, in vitro or in vivo fertilisation, in vivo or in vitro development and the different hormonal stimulation protocols used for obtaining the oocytes. All the genes analysed, their functions and abbreviations used in the present study are described in Table 2 and Fig. 1.
Expression of mitochondrial-encoded genes
The expression patterns of representative mitochondrial genes that comprise Complexes I–IV of the electron transport chain were characterised to evaluate the relationship between nuclear-encoded and mitochondrial-encoded transcripts (Figs 2–4). Maternally derived transcripts were detected for all the genes analysed: MT-ND1, MT-ND2, MT-ND6 and NDUFS1 (Fig. 2), SDHA, SDHB, SDHC (Fig. 3), MT-CO1, MT-CO2 and MT-CYTB (Fig. 4). MT-ND1 showed no significant variation throughout development (Fig. 2). MT-ND2 decreased significantly from the GV to MII stage, steadily decreasing throughout development, with significantly lower levels detected at the hatched blastocyst stage (Fig. 2). MT-ND6 decreased significantly from the GV to pronuclear (PN) stage, increased significantly at the eight-cell stage and again at the morula stage, remaining high through to the hatched blastocyst stage (Fig. 2). NDUFS1 mRNA expression decreased significantly following fertilisation, increasing at the two-cell stage and remaining high thereafter (Fig. 2). The expression of Complex II succinate dehydrogenases A and B mRNAs showed a significant reduction during oocyte maturation (Fig. 3). A gradual increase in SDHA mRNA was observed, with mRNA levels significantly higher at the eight-cell stage compared with PN embryos and significantly higher at the morula stage compared with two-cell embryos. The mRNA expression of SDHB increased significantly at the morula stage compared with PN, two- and eight-cell embryos, with transcript levels maintained through blastocyst development (Fig. 3). Significant differences in the expression of SDHC were detected between GV-stage oocytes and eight-cell and morula embryos, as well as between two- and eight-cell embryos (Fig. 3). Cytochrome b (MT-CYB; Complex III) was present in GV-stage oocytes, with mRNA expression increasing gradually through the eight-cell stage; this expression was partially sensitive to α-amanitin (Fig. 4). The expression of mitochondrially encoded cytochrome c oxidase subunit 1 (MT-CO1; Complex IV) mRNA was minimal in the GV-stage oocyte, increasing significantly at the two-cell stage through to the eight-cell stage, after which a significant decrease in expression was observed at the morula stage. MT-CO1 expression remained reduced through blastocyst formation. The expression of MT-CO1 was sensitive to α-amanitin (Fig. 4). The cytochrome c oxidase subunit 2 (MT-CO2) probe yielded strong hybridisation signals in oocytes and embryos of all stages, with a slight decrease in expression during the later cleavage stages (Fig. 4).
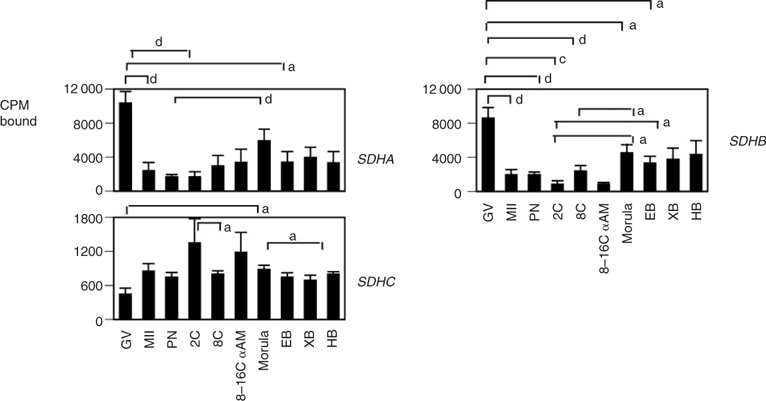
|

|
Expression of nuclear-encoded genes involved in mitochondrial transcription
The developmental expression patterns of key mitochondrial transcription regulators were analysed to determine the molecular events controlling mitochondrial transcription. Expression of the key components that regulate mitochondrial transcription was variable. The early stages of oocyte and embryo development were characterised by low levels of POLRMT, TFAM, TFB1M and TFB2M expression (Fig. 5). POLRMT mRNA expression increased progressively from the eight-cell stage to the blastocyst stage (Fig. 5). The TFB2M probe yielded low hybridisation signals, but transiently elevated expression was observed at the two-cell stage (Fig. 5). TFB1M expression increased steadily from the two-cell stage through the morula stage, with significantly higher levels at the morula and expanded blastocyst stages compared with MII oocytes (P = 0.014 and 0.02, respectively), PN zygotes (P = 0.02 and 0.001, respectively) and two-cell embryos (P = 0.03 and 0.01, respectively). This expression decreased during blastocyst development (Fig. 5), with significantly lower levels at the hatched blastocyst stage compared with the expanded blastocyst stage. TFAM expression was undetectable at the MII and two-cell stages. Transcriptional induction of the TFAM gene was observed at the eight-cell stage, with continued expression thereafter (Fig. 5). The mRNA encoding mitochondrial transcription termination factor (MTERF) was barely detected and expression fluctuated throughout development (Fig. 5).

|
The expression of the regulatory components of mitochondrial transcription is modulated by other nuclear-encoded factors, including NRF1 and NRF2 (also known as GA-binding protein; GABP). The expression of the GABPB (NRF2 β2-subunit) mRNA appeared significantly more abundant than that of NRF1, based on the overall strength of hybridisation signals (Fig. 5). GABPB mRNA was maternally expressed, decreased significantly from the GV to MII stage and increased at the PN stage. Expression of GABPB at the eight-cell stage was largely α-amanitin insensitive, indicating that the early expression of this mRNA was mostly due to maternal transcripts (Fig. 5). In comparison, NRF1 mRNA was not maternally expressed, but a transient α-amanitin-sensitive elevation in expression was seen at the eight-cell stage (Fig. 5). The mRNA encoding mitochondrial translation initiation factor MTIF2 was maternally expressed, first detected at the GV stage and declined significantly at the morula and early blastocyst stages (Fig. 5). An increase in MTIF2 mRNA was observed at the two-cell stage, although this was only significantly higher than the level of MTIF2 at the morula stage. MTIF3 mRNA was detected at a high level in the GV oocyte and at much lower levels at all other stages (Fig. 5).
Expression of nuclear-encoded genes involved in mitochondrial replication
The regulatory components involved in mitochondrial replication were examined to determine whether replication events could be detected at specific stages of development. Transcripts for POLG, SSBP1 and PEO1 (also known as TWINKLE) were maternally expressed. An increase in POLG was observed during oocyte maturation, followed by a gradual reduction with development to the PN and two-cell stages (Fig. 6). SSBP1 mRNA was predominantly expressed in embryos after the eight-cell stage. Maternal transcripts for PEO1 helicase were also detected, increasing significantly at the two-cell stage compared with maternal levels, decreasing significantly to the eight-cell stage, where expression remained α-amanitin insensitive. A further significant decrease in PEO1 mRNA levels occurred at the morula stage, with transcript levels remaining low through the hatched blastocyst stage (Fig. 6).

|
Effects of culture on expression of mitochondrial and nuclear transcription factor genes
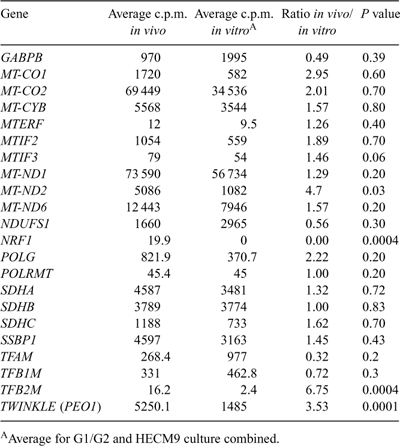
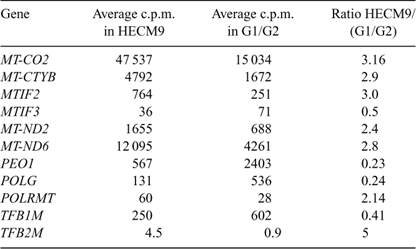
In vitro culture is known to affect gene expression patterns in preimplantation embryos (Niemann and Wrenzycki 2000), suggesting that the expression of genes regulating mitochondrial transcription and replication may be affected by culture in rhesus preimplantation blastocysts. A direct comparison between hatched blastocysts derived in vivo with those cultured in either G1/G2 or HECM9 revealed effects of culture environment on gene expression patterns (Table 3). NRF1 was detected at low levels in in vivo-derived blastocysts, but was undetectable in cultured blastocysts (P = 0.0004). Similarly, MT-ND2, TFB2M and PEO1 mRNA expression was significantly reduced in cultured embryos compared with their in vivo-derived counterparts. There was a trend for other transcripts to be affected by culture (MT-CO1, MT-CO2, POLG, TFAM), although these differences did not reach statistical significance. Differences in gene expression were also observed between G1/G2- and HECM9-cultured embryos for four mRNAs (Table 4). A higher level of expression of MT-CO2, MT-CYTB, MT-ND2 and MT-ND6 mRNAs was observed in hatched blastocysts cultured in HECM9 compared with those cultured in G1/G2. The expression of POLRMT and TFB2M mRNA was reduced in hatched blastocysts following culture using G1/G2, whereas TFB1M expression was higher in embryos cultured in G1/G2. POLG and PEO1 mRNA levels were reduced in hatched blastocysts cultured in HECM9 compared with those cultured in G1/G2. MTIF2 expression was lower in G1/G2-cultured blastocysts, whereas MTIF3 mRNA levels were lower in HECM9-cultured blastocysts. No other alterations in genes involved in mitochondrial transcription were detectable as a result of differences in the composition of the in vitro culture media used.
|
|
Effect of hormonal stimulation on the expression of mitochondrial genes and their transcription factors
Previous studies revealed that many maternal mRNAs become precociously degraded during development to the two-cell stage in embryos derived by in vitro maturation and fertilisation of oocytes isolated from NS females (Zheng et al. 2005; Mtango and Latham 2007, 2008). Given the importance of mitochondrial biogenesis and the trend for significant contribution of maternally encoded mRNAs to developmental patterns of expression outlined above, the expression of these mRNAs was compared between embryos that developed from NS oocytes and oocytes from FSH- or FSH+hCG-stimulated females (low, intermediate, and high developmental potential, respectively). Nine mRNAs displayed reduced expression at the two-cell stage in embryos developed from oocytes from NS females (Fig. 7). One of the nine mRNAs affected (PEO1) is also reduced in two-cell stage embryos from FSH-stimulated oocytes. Of the nine mRNAs affected, five (MT-CO1, MT-CYB, MT-ND1, MT-ND6, NDUFS1) pass the more stringent significance level of P < 0.016.
Discussion
The mitochondrion constitutes a critical component of the oocyte, providing for metabolic ATP-generating demands, as well as playing key regulatory roles in processes such as signal transduction and homeostasis. The data presented here reveal, for the first time, detailed temporal patterns of expression of mRNAs related to mitochondrial biogenesis in rhesus monkey oocytes and embryos. Persistent expression of maternally encoded mRNAs, combined with transcriptional activation and mRNA accumulation for many of these genes at the eight-cell stage, around the time of embryonic genome activation, was observed. The data also reveal that the expression of these mRNAs is greatly altered in embryos of reduced developmental potential, with the abundance of many maternally encoded mRNAs being precociously diminished. Finally, the data reveal a striking effect of embryo culture and specific media formulations on the expression of some of these mRNAs, including a deficiency in the expression of key transcriptional regulators.
Increasing expression, beginning at the eight-cell or morula stage, with evidence of α-amanitin sensitivity, was observed for several mRNAs examined (e.g. MT-ND6, SDHB, MT-CO1, NRF1, TFAM, POLG, POLRMT and TFB1M). Similar patterns have been observed for these transcripts in mouse, sheep and bovine embryos (May-Panloup et al. 2005; Thundathil et al. 2005; Bowles et al. 2007). However, several of the mRNAs examined (e.g. MT-ND1, MT-ND2, MT-CO1, MT-CYB, MTIF2 and TFB2) displayed predominantly maternally inherited expression that persisted through the eight-cell stage, followed by diminished expression thereafter. Other mRNAs were expressed both maternally and embryonically, with persistent expression (e.g. NDUFS1, SDHA, SDHC, MT-CO2, MTERF) during the period from the eight-cell to blastocyst stages. Collectively, these hybridisation patterns indicate that some genes are transcriptionally activated and direct increased accumulation of their mRNAs at the eight-cell stage, whereas others are either poorly transcribed at later stages or are transcribed at lower levels to direct more constant mRNA abundances.
Mitochondrial transcription requires cooperation between mitochondrial RNA polymerase (POLRMT), mitochondrial transcription factor A (TFAM) and at least one of the transcription factors TFB1M or TFB2M (Falkenberg et al. 2002). In addition to its role in mitochondrial transcription, TFAM is also required for replication of mtDNA (Kang and Hamasaki 2005). TFAM binds to and stabilises DNA, mediating mtDNA maintenance, and is therefore a crucial component for maintaining mtDNA copy number and respiratory chain activity (Larsson et al. 1998). Low or absent expression of TFAM mRNA during the early cleavage stages has been reported in the bovine (May-Panloup et al. 2005) and sheep (Bowles et al. 2007), with transcripts appearing at the morula stage and increasing at the blastocyst stage. A similar pattern of expression was observed during mouse embryo development (Thundathil et al. 2005) and is consistent with observations in the present study, where rhesus macaque embryos expressed TFAM at undetectable, or very low levels, during early development, increasing at the eight-cell stage with embryonic genome activation. These results are consistent with an increase in mitochondrial biogenesis as development proceeds, and possibly coincides with an increasing requirement for ATP (Leese 1995).
Both TFB1M and TFB2M cooperate with TFAM and POLRMT to initiate transcription (Falkenberg et al. 2002). The expression levels of TFB1M and TFB2M were relatively similar in the mouse (Thundathil et al. 2005); however, TFB2M expression in the rhesus was minimally detectable in the present study. Although this may imply that TFB1M plays a more significant role in the regulation of mitochondrial transcription in the non-human primate embryo, the expression of TFB2M was significantly reduced (by almost sevenfold) in in vitro-cultured blastocysts compared with in vivo-derived embryos (Table 3). This suggests that in vitro-cultured monkey embryos must rely on TFAM binding with TFB1M, a less efficient cooperation (Falkenberg et al. 2002), to enable mtDNA transcription to occur and may have less flexibility in regulating mitochondrial transcription. Likewise, POLRMT expression levels were low during rhesus macaque oocyte maturation and early embryo development. Low levels of the components of mitochondrial transcription may be insufficient, which may impact mitochondrial transcription and, in turn, lead to metabolic deficiencies and poor developmental outcomes. TFB1M mRNA expression in the rhesus was relatively low, but consistent through to the two-cell stage, increasing from the eight-cell to morula stages and decreasing with blastocyst formation (Fig. 5). This also contrasts with TFB1M mRNA expression in the mouse, which decreases significantly at the two-cell stage, increasing significantly through to the blastocyst stage (Thundathil et al. 2005). Whether the low levels of expression of TFB1M in the rhesus monkey are sufficient to compensate for the absence of TFB2M to enable continued and appropriate levels of mtDNA transcription requires further investigation.
The culture environment exerted a significant effect on mitochondrial gene expression. Reduced expression of several mRNAs was observed in cultured embryos compared with flushed embryos, indicating that culture may compromise energy metabolism. The control of energy metabolism is based on the efficiency of energy transduction by cytochrome c oxidase (Complex IV). Cytochrome oxidase mRNAs displayed highly divergent patterns of expression between rhesus monkey embryos and those reported previously in bovine and mouse (May-Panloup et al. 2005; Thundathil et al. 2005). Significantly, a twofold decrease in MT-CO1 and a threefold reduction in MT-CO2 expression was observed in cultured rhesus macaque hatched blastocysts compared with in vivo-derived blastocysts, suggesting that the proton pumping efficiency of the cytochrome c oxidase complex may be deficient in cultured rhesus monkey embryos and may contribute to the reduced developmental potential of in vitro-matured oocytes. In addition, whereas HECM9 medium supported an overall higher expression level for many mtDNA-encoded mRNAs compared with G1/G2 medium, higher levels of expression of replication factors PEO1 and POLG were observed in the latter. These differences may be attributable to alterations in cellular redox status (Harvey et al. 2002), such that suboptimal culture media may directly perturb mitochondrial gene expression, as well as the nuclear components regulating mitochondrial biogenesis.
May-Panloup et al. (2005) reported the mtDNA content of bovine oocytes and embryos based on the expression of MT-ND2. MT-ND2 expression in the bovine followed a similar pattern to that observed by Thundathil et al. (2005) in the mouse, with transcript levels decreasing at the two-cell stage and increasing at the morula stage. However, these profiles contrast with the continual decline in MT-ND2 levels in rhesus macaque embryos observed in the present study (Fig. 2). Moreover, a significant reduction in MT-ND2 mRNA levels following in vitro culture in primate blastocysts was observed compared with their in vivo-derived counterparts (Table 3). Conversely, the expression pattern of MT-ND6 in the monkey was similar to that reported for the mouse, with the number of transcripts declining slightly during early development from the oocyte, then increasing at the morula stage (Fig. 2, Thundathil et al. 2005). This suggests that these transcripts are inappropriate markers of mtDNA copy number, particularly for preimplantation embryos, where in vitro culture conditions may alter the expression of mitochondrial transcripts.
TFB1M and TFB2M, which cooperate with TFAM and POLRMT to initiate transcription, are regulated by NRF1 and NRF2 (GABPB; Gleyzer et al. 2005; Scarpulla 2006). Absent or very low NRF1 mRNA expression in cultured embryos may impede mitochondrial transcription, in addition to altering the relative abundance of TFB1M and TFB2M, thereby having a downstream effect on oxidative phosphorylation and potentially limiting the viability of in vitro-produced rhesus macaque embryos. Disruption of NRF1 expression in knockout mouse blastocysts has been shown to impact mtDNA copy number (Huo and Scarpulla 2001). These results further suggest that the initial burst in expression of genes related to mitochondrial biogenesis, coupled with maternally inherited mRNAs, supports mitochondrial biogenesis through to the blastocyst stage, but that the ability to undergo mitochondrial biogenesis may be limited owing to lower embryo quality resulting from culture conditions and oocyte and/or embryo heterogeneity.
Mitochondrial transcription produces the RNA primers required for the initiation of mtDNA replication at the origin of H-strand DNA replication (OH; Clayton 1991). It seems logical to assume that, if mitochondrial transcription is significantly affected by in vitro culture, the initiation of mitochondrial replication and subsequent processing may also be affected. mtDNA is replicated by POLG and replication factors, including mitochondrial single-stranded DNA binding protein and PEO1 (Twinkle helicase; Falkenberg et al. 2007), the three of which form the minimal replisome required for mtDNA replication to occur. PEO1 expression was significantly reduced (threefold) in cultured embryos compared with in vivo-derived blastocysts (Table 3). The ability to unwind mtDNA is an essential process required for replication by POLG (Korhonen et al. 2003). Aberrations in PEO1 expression are associated with a higher incidence of mtDNA mutation (Tyynismaa et al. 2005) and a rapid decrease in mtDNA copy number (Tyynismaa et al. 2004). Furthermore, a twofold decrease in POLG expression was observed between cultured and in vivo-derived embryos. Although this overall difference was not significant, the data suggest that deficiencies in mitochondrial replication may exist in cultured rhesus embryos.
POLG is the only known mitochondrial polymerase and is thereby responsible for mtDNA replication and repair functions (for a review, see Kaguni 2004). A reduction in POLG levels or mutations in POLG have been associated with mtDNA mutations. Although the common deletion has been detected in rhesus oocytes and embryos (Gibson et al. 2005), it is unclear the extent to which mtDNA mutations are present in rhesus embryos. Mutation of either POLG or PEO1 also results in reduced mtDNA copy number as a result of replication stalling (Wanrooij et al. 2007). A reduction in both PEO1 and POLG may be indicative that replication stalling is occurring in in vitro-cultured embryos. Evidence in rhesus macaque oocytes suggests that replication stalling may also occur during oocyte maturation (Gibson et al. 2005). Mutations in PEO1 have also been associated with progressive cytochrome c oxidase deficiency in mouse neurons (Tyynismaa et al. 2005). Notably, a two- to threefold reduction in MT-CO1 and MT-CO2, respectively, was observed in in vitro-cultured rhesus embryos compared with in vivo-derived embryos. Therefore, alterations in the expression of the mitochondrial replication machinery may indicate a reduction in mtDNA replication capacity. The aberrant expression of factors regulating mtDNA transcription and/or replication may adversely affect embryo development through altered nuclear–mitochondrial cross-talk, and subsequently altered ATP generation through the electron transport chain, associated with a depletion of mtDNA copy number. Failure of TFAM, TFB1M and TFB2M to coordinate transcription, or of TFAM and POLG to coordinate replication, has been shown to impact post-implantation development. Homozygous knockout mice for Tfam and Polg fail to survive, as reported by Hance et al. (2005) and Larsson et al. (1998), respectively, whereas heterozygotes for Tfam exhibit reduced mtDNA copy number and respiratory chain deficiencies. Polg heterozygotes exhibit severe cardiomyopathies and slight reductions in mtDNA copy number. Deficiencies in regulating nuclear–mitochondrial cross-talk and the susceptibility of mitochondrial biogenesis to in vitro maturation and culture, as observed in the present study, may underlie the poor success rates of human assisted reproductive technologies, particularly in terms of ATP production and acquiring the minimum mitochondrial threshold required for continued development (Reynier et al. 2001).
Defective nuclear–cytoplasmic communication may also contribute to an inability to detect clear replication events. It is unclear whether additional replication events occur during rhesus monkey preimplantation development to support post-implantation development. A non-statistically significant increase in POLG was observed from the GV to MII stage, likely coinciding with the previously reported replication event during the late stages of oocyte maturation in the monkey (Gibson et al. 2005), which is also observed in the pig (Spikings et al. 2007). A window of mtDNA replication has also been detected post-fertilisation in mice (McConnell and Petrie 2004). A similar small, non-significant increase in POLG was observed in the present study around the time of embryonic genome activation at the eight-cell stage, coinciding with an increase in TFAM and SSBP1 expression. Significantly, Kameyama et al. (2007) reported that in vitro culture induced an increase in mtDNA copy number at the eight-cell stage, supporting the hypothesis that suboptimal culture conditions can have a significant effect on mitochondrial biogenesis.
The effects of hormonal stimulation and in vitro maturation on mitochondrial gene expression in two-cell embryos are striking. Oocytes matured in vitro from NS rhesus females display poor developmental competence (Zheng et al. 2005) compared with ‘in vivo-matured’ oocytes. In vitro maturation of oocytes from FSH-primed females yields intermediate-quality oocytes (Schramm and Bavister 1994). Data reveal that, like many other maternally encoded mRNAs, some maternally encoded mRNAs related to mitochondrial biogenesis are eliminated precociously and are thus deficient in embryos developed from NS oocytes and matured in vitro. Hence, defects in mitochondrial biogenesis may contribute to the reduced developmental potentials of these embryos, manifesting as an inability to regulate electron transport.
Previous studies have revealed significant differences between mouse and human or non-human primate embryonic patterns of gene expression (Vassena et al. 2005; Zheng et al. 2005; Mtango and Latham 2007, 2008), highlighting the value of a non-human primate model for detailed studies of developmental transitions and developmental mechanisms that may drive human embryogenesis. In addition, critical comparisons to understand the molecular basis of defective embryogenesis require the expenditure of high-quality oocytes and embryos for molecular analysis, which is precluded for the human by legal and ethical considerations. The PREGER resource was developed as a system of more than 170 cDNA libraries that can be used for basic studies of gene expression patterns. The data presented here, along with other published data, collectively illustrate the dynamic nature of gene regulation related to mitochondrial biogenesis, as well as a clear propensity for the expression of these genes to be adversely affected by changes in oocyte maturation and embryo culture protocols. Investigations are ongoing to determine whether changes in mitochondrial replication and transcription factor expression influence mitochondrial function, mtDNA mutation rates or mtDNA copy number in oocytes or embryos. The sensitivity of this group of genes to such factors highlights the need for additional studies in human and non-human primate model species in order to determine in detail how mitochondrial biogenesis can be affected by oocyte and embryo manipulation protocols and whether this may affect physiological functions in the resulting progeny.
Acknowledgements
The authors thank Bela Patel, Malgorzata McMenamin, Judy Procknow and Ann Marie Paprocki for their technical assistance. The authors also thank R. Dee Schramm for his contribution to the development of the PREGER resource. This work was supported by a research resource grant from the National Centers for Research Resources (RR15253) and CA95569 to K.L. and a National Centers for Research Resources grant (RR021881) to C.B.
Acton, B. M. , Jurisicova, A. , Jurisica, I. , and Casper, R. F. (2004). Alterations in mitochondrial membrane potential during preimplantation stages of mouse and human embryo development. Mol. Hum. Reprod. 10, 23–32.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
Lee, D. Y. , and Clayton, D. A. (1996). Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J. Biol. Chem. 271, 24 262–24 269.
| PubMed |
Scarpulla, R. C. (2006). Nuclear control of respiratory gene expression in mammalian cells. J. Cell. Biochem. 97, 673–683.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Scarpulla, R. C. (2008). Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88, 611–638.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schramm, R. D. , and Bavister, B. D. (1994). Follicle-stimulating hormone priming of rhesus monkeys enhances meiotic and developmental competence of oocytes matured in vitro. Biol. Reprod. 51, 904–912.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schramm, R. D. , and Bavister, B. D. (1995). Effects of granulosa cells and gonadotrophins on meiotic and developmental competence of oocytes in vitro in non-stimulated rhesus monkeys. Hum. Reprod. 10, 887–895.
| PubMed |

Schramm, R. D. , and Bavister, B. D. (1996). Granulosa cells from follicle stimulating hormone-primed monkeys enhance the development competence of in-vitro-matured oocytes from non-stimulated rhesus monkeys. Hum. Reprod. 11, 1698–1702.
| PubMed |

Shadel, G. S. , and Clayton, D. A. (1997). Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66, 409–435.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Smith, L. C. , and Alcivar, A. A. (1993). Cytoplasmic inheritance and its effects on development and performance. J. Reprod. Fertil. Suppl. 48, 31–43.
| PubMed |

Spikings, E. C. , Alderson, J. , and John, J. C. (2007). Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biol. Reprod. 76, 327–335.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Squirrell, J. M. , Lane, M. , and Bavister, B. D. (2001). Altering intracellular pH disrupts development and cellular organization in preimplantation hamster embryos. Biol. Reprod. 64, 1845–1854.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Squirrell, J. M. , Schramm, R. D. , Paprocki, A. M. , Wokosin, D. L. , and Bavister, B. D. (2003). Imaging mitochondrial organization in living primate oocytes and embryos using multiphoton microscopy. Microsc. Microanal. 9, 190–201.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Steuerwald, N. , Barritt, J. A. , Adler, R. , Malter, H. , Schimmel, T. , Cohen, J. , and Brenner, C. A. (2000a). Quantification of mtDNA in single oocytes, polar bodies and subcellular components by real-time rapid cycle fluorescence monitored PCR. Zygote 8, 209–215.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Steuerwald, N. , Cohen, J. , Herrera, R. J. , and Brenner, C. A. (2000b). Quantification of mRNA in single oocytes and embryos by real-time rapid cycle fluorescence monitored RT-PCR. Mol. Hum. Reprod. 6, 448–453.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Sturmey, R. G. , and Leese, H. J. (2003). Energy metabolism in pig oocytes and early embryos. Reproduction 126, 197–204.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Taylor, K. D. , and Piko, L. (1995). Mitochondrial biogenesis in early mouse embryos: expression of the mRNAs for subunits IV, Vb, and VIIc of cytochrome c oxidase and subunit 9 (P1) of H(+)-ATP synthase. Mol. Reprod. Dev. 40, 29–35.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Thundathil, J. , Filion, F. , and Smith, L. C. (2005). Molecular control of mitochondrial function in preimplantation mouse embryos. Mol. Reprod. Dev. 71, 405–413.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Trimarchi, J. R. , Liu, L. , Porterfield, D. M. , Smith, P. J. , and Keefe, D. L. (2000). Oxidative phosphorylation-dependent and -independent oxygen consumption by individual preimplantation mouse embryos. Biol. Reprod. 62, 1866–1874.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tyynismaa, H. , Sembongi, H. , Bokori-Brown, M. , Granycome, C. , Ashley, N. , Poulton, J. , Jalanko, A. , Spelbrink, J. N. , Holt, I. J. , and Suomalainen, A. (2004). Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 13, 3219–3227.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tyynismaa, H. , Mjosund, K. P. , Wanrooij, S. , Lappalainen, I. , Ylikallio, E. , Jalanko, A. , Spelbrink, J. N. , Paetau, A. , and Suomalainen, A. (2005). Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl Acad. Sci. USA 102, 17 687–17 692.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Van Blerkom, J. (2004). Mitochondria in human oogenesis and preimplantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction 128, 269–280.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Van Blerkom, J. , Bell, H. , and Weipz, D. (1990). Cellular and developmental biological aspects of bovine meiotic maturation, fertilization, and preimplantation embryogenesis in vitro. J. Electron Microsc. Tech. 16, 298–323.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Van Blerkom, J. , Davis, P. , and Alexander, S. (2000). Differential mitochondrial distribution in human pronuclear embryos leads to disproportionate inheritance between blastomeres: relationship to microtubular organization, ATP content and competence. Hum. Reprod. 15, 2621–2633.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Vassena, R. , Dee Schramm, R. , and Latham, K. E. (2005). Species-dependent expression patterns of DNA methyltransferase genes in mammalian oocytes and preimplantation embryos. Mol. Reprod. Dev. 72, 430–436.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Virbasius, J. V. , and Scarpulla, R. C. (1994). Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl Acad. Sci. USA 91, 1309–1313.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wanrooij, S. , Goffart, S. , Pohjoismaki, J. L. , Yasukawa, T. , and Spelbrink, J. N. (2007). Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 35, 3238–3251.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wilding, M. , Dale, B. , Marino, M. , di Matteo, L. , Alviggi, C. , Pisaturo, M. L. , Lombardi, L. , and De Placido, G. (2001). Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum. Reprod. 16, 909–917.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Zheng, P. , Patel, B. , McMenamin, M. , Reddy, S. E. , Paprocki, A. M. , Schramm, R. D. , and Latham, K. E. (2004). The primate embryo gene expression resource: a novel resource to facilitate rapid analysis of gene expression patterns in non-human primate oocytes and preimplantation stage embryos. Biol. Reprod. 70, 1411–1418.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Zheng, P. , Patel, B. , McMenamin, M. , Moran, E. , Paprocki, A. M. , Kihara, M. , Schramm, R. D. , and Latham, K. E. (2005). Effects of follicle size and oocyte maturation conditions on maternal messenger RNA regulation and gene expression in rhesus monkey oocytes and embryos. Biol. Reprod. 72, 890–897.
| Crossref | GoogleScholarGoogle Scholar | PubMed |





