399 SPERMATOZOA-MEDIATED GENE TRANSFER IS INFLUENCED BY SIZE, STRUCTURE, AND CONCENTRATION OF EXOGENOUS DNA
W. B. Feitosa A , M. P. Milazzotto A , E. A. L. Martins B , A. M. Rocha A , J. L. Avanzo C , J. A. Visintin A and M. E. O. A. Assumpção AA Department of Animal Reproduction, FMVZ, São Paulo University, São Paulo, SP, Brazil
B Biotechnology Center, Butantan Institute, São Paulo, SP, Brazil
C Department of Animal Pathology, FMVZ, São Paulo University, São Paulo, SP, Brazil
Reproduction, Fertility and Development 19(1) 315-315 https://doi.org/10.1071/RDv19n1Ab399
Submitted: 12 October 2006 Accepted: 12 October 2006 Published: 12 December 2006
Abstract
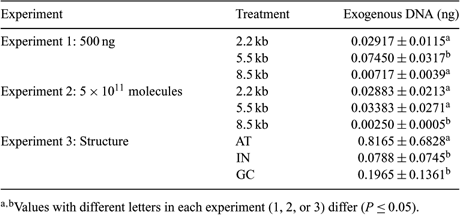
Mammalian sperm cells have a spontaneous ability to take up exogenous DNA in a process regulated by specific protein. However, little is known about the effect of exogenous DNA on sperm cell association. The aim of this work was to evaluate the effect of size, concentration, and structure of exogenous DNA sequence in apoptosis and oncosis induction and the association with spermatozoa. To evaluate the effect of size and concentration of exogenous DNA, sequences of 2.2, 5.5, and 8.5 kb were used at the concentrations of 500 ng (Experiment 1) or 5 × 1011 molecules (Experiment 2). To assess the effect of structure (Experiment 3), 3 sequences of approximately 500 bp were used, containing 65.7% of AT (AT sequence), 46.4% of AT (IN sequence), and 38.6% of AT (GC sequence). To evaluate apoptosis and oncosis in all treatments, sperm cells were incubated with exogenous DNA for 1 h, stained with 2 µL of Yo-Pro (100 µM) for 20 min, and 10 µL of propidium iodide (6 µM) for 10 min, and then analyzed by flow cytometry. To evaluate the transfection rates, sperm cells, after incubation, were separated from non-associated exogenous DNA by 2.5% gel electrophoresis. DNA (genomic and exogenous) was extracted and association rates obtained by real-time PCR. Data were submitted to ANOVA and compared by Tukey's test (P ≤ 0.05). The number of viable, apoptotic, oncosis, or in initial apoptosis/late oncosis spermatozoa incubated with different concentrations or sequences of exogenous DNA did not differ among groups. In Experiment 1, a higher DNA association with sperm cells was observed in the 5.5 kb sequence in comparison with the 2.2 and 8.5 kb. In Experiment 2, the 8.5 kb sequence had the lower association with the spermatozoa in comparison with the 2.2 kb and 5.5 kb. In Experiment 3, there was a higher interaction of the AT sequence when compared to IN and GC sequences. Reference data suggest that big sequences of exogenous DNA have advantages in the interaction with spermatozoa. The present work is the first one to show that the advantage of the big sequences on interaction became a disadvantage in relation to the ability of these cells to internalize this DNA.
|
Financial support was provided by FAPESP 03/10234-7 and 03/07456-8.


