180 IDENTIFICATION AND QUANTIFICATION OF DIFFERENTIALLY REPRESENTED TRANSCRIPTS IN PREIMPLANTATION BOVINE EMBRYOS
C. E. McHughes A , G. K. Springer A , L. D. Spate A , R. Li A , R. J. Woods A , M. P. Green A , S. W. Korte A , C. N. Murphy A , J. A. Green A and R. S. Prather AUniversity of Missouri, Columbia, MO, USA
Reproduction, Fertility and Development 20(1) 169-170 https://doi.org/10.1071/RDv20n1Ab180
Published: 12 December 2007
Abstract
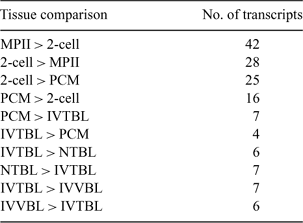
Identification of transcripts that are present at key development stages of preimplantation embryos is critical for a better understanding of early embryogenesis. To that end, this project had two goals. The first was to characterize the relative abundance of multiple transcripts during several developmental stages, including metaphase II-stage oocytes (MPII), and 2-cell-stage (2-cell), precompact morula (PCM), and in vitro-produced blastocyst-stage (IVTBL) embryos. The second was to characterize differences in the relative abundance of transcripts present in in vivo- (IVVBL), in vitro-, and nuclear transfer-produced (NTBL) blastocysts. It was our hypothesis that the identification of differentially represented transcripts from these stages would reveal not only developmentally important genes, but also genes that might be aberrantly expressed due to embryo production techniques. Individual clusters from a large bovine EST project (http://genome.rnet.missouri.edu/Bovine/), which focused on female reproductive tissues and embryos, were compared using Fisher's exact test weighted by number of transcripts per tissue by gene (SAS PROC FREQ; SAS Institute, Inc., Cary, NC, USA). Of the 3144 transcripts that were present during embryogenesis, 125 were found to be differentially represented (P < 0.01) in at least one pairwise comparison (Table 1). Some transcripts found to increase in representation from the MPII to the 2-cell stage include protein kinases, PRKACA and CKS1, as well as the metabolism-related gene, PTTG1. These same transcripts were also found to decrease in representation from the 2-cell to the PCM stage. RPL15 (translation) and FTH1 (immune function) were both more highly represented in the PCM than in the 2-cell stage. From PCM to IVTBL, we saw an increase in RPS11, another translation-related transcript. When comparing blastocyst-stage embryos from different production techniques, several transcripts involved in energy production (e.g., COX7B and COX8A) were found to be more highly represented in the NTBL than in the IVTBL. COX8A was also more highly represented in the IVVBL than in the IVTBL. By investigating these differentially represented transcripts, we will be able to better understand the developmental implications of embryo manipulation. We may also be able to better develop reproductive technologies that lead to in vitro- and nuclear transfer-derived embryos which more closely follow a normal program of development.
|


