Phylogeny of the holly grevilleas (Proteaceae) based on nuclear ribosomal and chloroplast DNA
Gareth D. Holmes A B C , Trisha L. Downing A , Elizabeth A. James A C , Mark J. Blacket D , Ary A. Hoffmann B and Michael J. Bayly A EA School of Botany, The University of Melbourne, Parkville, Vic. 3010, Australia.
B Department of Genetics, Bio21 Molecular Science and Biotechnology Institute, 30 Flemington Road, The University of Melbourne, Vic. 3010, Australia.
C National Herbarium of Victoria, Royal Botanic Gardens Melbourne, Birdwood Avenue, South Yarra, Vic. 3141, Australia.
D Department of Environment and Primary Industries, AgriBio, La Trobe University, 5 Ring Road, Bundoora, Vic. 3083, Australia.
E Corresponding author. Email: mbayly@unimelb.edu.au
Australian Systematic Botany 27(1) 56-77 https://doi.org/10.1071/SB13045
Submitted: 1 November 2013 Accepted: 8 May 2014 Published: 30 June 2014
Abstract
The holly grevilleas are an informal grouping of 15 species (19 taxa) of woody shrubs from south-eastern Australia, with a centre of distribution in central to western Victoria. Many of the species are narrowly endemic. The present study is the first molecular-phylogenetic analysis of the group, with the aim of providing an evolutionary framework for assessing species-level taxonomy and conservation priorities. Analyses using the nrDNA internal transcribed spacer (ITS) regions were complicated by the presence of divergent paralogues, including inferred pseudogenes; analyses restricted to presumed orthologous, functional ITS sequences were uninformative. Combined analyses of three chloroplast intergenic spacers (trnQ–5′rps16, trnL–trnF and rpoB–trnC) strongly support the monophyly of a core group of 16 taxa (the ‘southern holly grevilleas’) from Victoria and South Australia. However, nodes outside this group are poorly resolved and poorly supported, and the relationships of taxa from New South Wales and eastern Victoria (the ‘northern holly grevilleas’) are unclear. Among the southern holly grevilleas, the following four distinct and partly sympatric cpDNA clades are identified: the ‘Grevillea ilicifolia’, ‘G. aquifolium’, ‘G. dryophylla’ and ‘G. repens’ clades, among which the earliest and most strongly supported divergence is that of the western-most ‘G. ilicifolia’ clade. Variation in cpDNA is incongruent with current species-level taxonomy, especially for G. aquifolium (polyphyletic), G. montis-cole (polyphyletic, but the two subspecies each monophyletic) and G. microstegia (nested in G. aquifolium). The effects of incomplete chloroplast lineage sorting, gene flow through hybridisation or introgression, and inappropriate taxonomy are possible explanations for this incongruence. The formal conservation listing for some species within the holly grevillea group requires re-evaluation.
Additional keywords: Grevillea aquifolium group, holly-leaved grevilleas, ITS, molecular phylogeny, Proteaceae, rDNA pseudogenes, rpoB–trnC, taxonomy, trnL–trnF, trnQ–5′rps16.
Introduction
Grevillea R.Br. ex Knight (Proteaceae) is a genus in which there is considerable taxonomic uncertainty. The latest treatment lists 362 recognised species, 357 of which are endemic to Australia (Makinson 2000), and a small number endemic to (or also found in) New Caledonia, Papua New Guinea and Sulawesi. Several new taxa have also been described since that publication (e.g. Downing et al. 2004). As currently defined, Grevillea is the third largest genus of flowering plants in Australia, after Acacia and Eucalyptus (George 1998).
The size of the genus Grevillea and the range of forms found within it complicate the application of formal infrageneric taxonomy and the delineation of some taxa using traditional taxonomic methods. Two informal treatments (McGillivray and Makinson 1993; Makinson 2000) and an extensive, horticulturally oriented taxonomic reworking with narrower species treatments (Olde and Marriott 1994, 1995a, 1995b) have been applied in recent times after the last formal treatment over a century ago (Bentham 1870). Despite the extensive work of these taxonomists, the relationships among species and the taxonomy of some sections of the group are still contentious. The current classification is predominantly based on comparative morphology, with some phylogenetic considerations, and divides the genus into 33 groups (Makinson 2000). However, there have been no phylogenetic analyses for the entire genus.
The holly-leaved grevilleas (or G. aquifolium group of species) are one group that warrants closer phylogenetic attention. This informal group of small woody shrubs is placed, along with 33 other species, in the Aspleniifolia–Hookeriana subgroup of the Pteridifolia group (Makinson 2000). It corresponds to Species 13–26 in Makinson (2000), Group 1.2.1.5 in the classification of McGillivray and Makinson (1993), part of Group 35 as described by Olde and Marriott (1994), and also closely to series Hybegynae of Bentham’s (1870) classification. The group has been defined as including ‘species with holly-like leaves and ‘toothbrush’ inflorescences, and…their close relatives’ (McGillivray and Makinson 1993). These features are potentially apomorphic within the genus, but have not been tested by phylogenetic analysis.
The holly grevilleas are distributed in south-eastern mainland Australia, and include 15 species (19 taxa). The group’s distribution is centred on central to western Victoria, but extends to the Eyre Peninsula, South Australia, to the west and to largely disjunct sites in southern New South Wales and near the Queensland–New South Wales border to the north-east (Fig. 1). Many taxa are narrowly endemic, especially in western Victoria (nine species, Fig. 1). Their evolutionary relationships and taxonomy are somewhat unclear because there is great phenotypic variability within some species, leading to several new species being recognised in recent decades (e.g. Molyneux 1975, 1985; Smith 1981, 1983). For example, G. ilicifolia was recently split into three species (two with subspecies) on the basis of leaf morphology (Downing et al. 2004), whereas G. williamsonii is no longer recognised, being considered a male-sterile morphological variant of G. aquifolium (James 2000, 2004; Walsh and Stasjic 2007). The species G. aquifolium has been the source of taxonomic contention, with many populations displaying distinct forms or ecological niches (e.g. Olde and Marriott 1995a). In all, 12 of the 19 recognised taxa were listed as Rare, Vulnerable or Endangered on the most recent ‘Rare or Threatened Australian Plants’ (ROTAP) list (Briggs and Leigh 1996) and six are currently considered vulnerable on the EPBC Act list of threatened flora (http://www.environment.gov.au/cgi-bin/sprat/public/publicthreatenedlist.pl?wanted=flora, accessed 17 July 2013). Several populations are known to be extinct or close to extinction (e.g. G. infecunda in south-eastern Melbourne, G. ilicifolia in New South Wales, G. aquifolium from near Portland) (Olde and Marriott 1995a). Two of the species in the group (G. infecunda and G. renwickiana) are thought to be sterile, displaying obligate clonal reproduction by ‘root-suckering’ (Kimpton et al. 2002; James and McDougall 2014).

|
The aim of the current study was to investigate relationships among the holly grevilleas by using DNA sequence data. The work was undertaken to provide an evolutionary framework for assessing species-level taxonomy and conservation priorities within the group. Given uncertainty surrounding higher-level relationships within Grevillea, and based on their morphological resemblance and informal classification, the holly grevilleas are considered a priori a cohesive group for phylogenetic study, even though their monophyly has not been tested by higher-level phylogenetic analysis.
Materials and methods
Taxon sampling
Leaf material (desiccated in silica gel for molecular analyses) and voucher herbarium specimens were obtained from each of the 15 recognised species (19 taxa) in the holly grevillea group. Where possible, more than one accession was collected for each ingroup taxon, particularly for those with disjunct or extensive geographical ranges and those with distinct morphological or ecological forms. In particular, sampling within G. aquifolium was targeted to ensure that the range of observed morphological variation across its geographic range was included in the analysis. Outgroup taxa included five species from the Pteridifolia group (Makinson 2000), namely three eastern members (G. acanthifolia, G. laurifolia, G. willisii) and one Western Australian member (G. dryandroides) of the Aspleniifolia–Hookeriana subgroup, and one Western Australian member of the Bipinnatifida subgroup (G. bipinnatifida). G. alpina (Linearifolia subgroup, Floribunda group sensu Makinson 2000), along with two species from the sister genus Hakea, were also included as outgroups for some analyses of internal transcribed spacer (ITS) sequences; these additional outgroups could not be used for analyses of cpDNA because sequence variation made alignment problematic. Samples were sourced from natural populations or from garden or nursery material grown from field-collected specimens (Table 1).
DNA isolation, polymerase chain reaction (PCR) and sequencing
DNA was isolated from ground, dried leaf tissue by using either a DNeasy Plant Mini Kit (QIAGEN, Valencia, CA, USA) following the manufacturer’s protocol, or at the Australian Genome Research Facility (AGRF), Adelaide, by using a Nucleospin Plant L extraction kit (Machery–Nagel, Düren, Germany), following the PL1 method outlined in the product manual (J. Chambers, AGRF Adelaide, pers. comm. 2009). All DNA isolates were stored at −20°C.
The nuclear rDNA ITS regions (ITS-1, 5.8S, ITS-2) and small segments of the flanking 18S and 26S ribosomal subunits were successfully amplified for a limited number of taxa in the study group using the primer pairs ITS5m–ITS4 (ITS5m, Sang et al. 1995; ITS4, White et al. 1990) or the new primers PrF (5′-GCGAGAAGTCCACTGAACC-3′)–PrR (5′-CTGAGGACGCTTCTACAGAC-3′). However, because of amplification specificity problems, difficulties sequencing through ITS-1 in the majority of taxa and the presence of paralogous ITS copies, we designed a primer specific to the putative functional ITS copy that excluded ITS-1 (PriA; 5′-GAACATCACAACGGAACGGG-3′) to be coupled with the reverse primers PrR or Pr2R (5′-GCCCGATTCTCAAGCTGG-3′). Subsequently, only the 5.8S, ITS-2 of the ITS and partial 26S subunit (hereafter referred to as ITS2) was amplified with these primers. The complementary strands for this DNA fragment were subsequently sequenced with the forward primer PriA, and PrR, Pr2R or ITS4 as a reverse primer. A summary of approaches attempted for amplifying the ITS is presented in appendix 1 of Holmes (2008).
The following PCR reagents and conditions proved successful for amplifying the putative functional ITS2 fragment: 30-μL reactions consisted of 1.0 U DNA polymerase (Immolase, Bioline, Alexandria, NSW), 200 μM dNTPs (Bioline), 2.0 mM MgCl2, 1 × PCR buffer (Immobuffer, Bioline), 0.5 μM of each primer, 0.3 μL 100 × bovine serum albumin (BSA) (New England BioLabs, Ipswich, MA, USA), 5% (v/v) dimethyl sulfoxide (DMSO), 0.5–1.5 μL genomic DNA (~10–50 ng) and dH20 to volume. PCR conditions consisted of an initial denaturation cycle at 95°C for 7 min, followed by 30 cycles of 30 s at 95°C, 45 s at 49°C and 45 s at 72°C. The last cycle was followed by an extension step of 3 min at 72°C. Reactions were performed using a GeneAmp 9700 (Perkin–Elmer, Waltham, MA, USA) or a Mastercycler gradient thermal cycler (Eppendorf, North Ryde, NSW).
After an initial screening of nucleotide variation across multiple samples within several chloroplast DNA regions (Holmes 2008; Downing 2012), three intergenic spacers from the large single-copy genome region (trnQ(UUG)–5′rps16, trnL(UAA)–trnF(GAA) and rpoB–trnC(GCA)) were amplified for all samples. The primers used for the trnQ–5′rps16 spacer were trnQ(UUG) and rps16x1 (Shaw et al. 2007). For the trnL–trnF spacer, the primers ‘e’ and ‘f’ from (Taberlet et al. 1991) were used. The trnC end of the rpoB–trnC spacer was amplified using the primer trnCGCAR (Shaw et al. 2005) and the new grevillea-specific internal primer BCif (TCGCAGGACAAAGAACAAAG); PCR amplification using the latter primer was successful for all sampled taxa, except G. dryandroides subsp. hirsuta, which was found to have a deletion overlapping the primer site.
For cpDNA amplification, several protocols were used initially, dependent on the DNA region of interest and availability of reagents (Holmes 2008). Most PCR reactions (25 µL) contained 0.2 µM of each forward and reverse primer, 2.5 mM of each dNTP, 0.1 µL of HotStarTaq DNA polymerase with 2.5 µL of 10 × PCR buffer (QIAGEN), 5–50 ng of genomic DNA, and dH20 to volume. For the rpoB–trnC and trnQ–5′rps16 intergenic spacers, reactions contained 2.0 mM MgCl2 and 0.5 µL of non-acetylated BSA (Fermentas, Vilnius, Lithuania), 0.1 µL of HotStarTaq DNA polymerase, 2.5 µL of 10 × PCR buffer (QIAGEN), 2.0 µL of DNA isolate and dH20 to volume.
CpDNA PCR amplifications were performed on an Eppendorf Mastercycler with the lid heated to 105°C. A ‘touch-down’ PCR protocol was employed for the trnL–trnF spacer. The PCR conditions were as follows: one cycle at 95°C for 15 min; five cycles of 95°C for 30 s, 64°C for 30 s (decreasing by 2.0°C each cycle), 72°C for 1 min; 30 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 1 min; a final extension of 72°C for 10 min. For the trnQ–5′rps16 and rpoB–trnC spacers, the PCR protocol was based on the ‘slow and cold’ ‘rpl16’ program of Shaw et al. (2007). PCR conditions consisted of one cycle at 95°C for 15 min; 30 cycles of 95°C for 1 min, 50°C for 1 min (increasing by 0.5°C each cycle), 65°C for 4 min; followed by a final extension of 65°C for 5 min.
After checking PCR amplification success, products were purified using a QIAquick purification kit (QIAGEN), Illustra GFX PCR DNA purification kit (GE Healthcare, Little Chalfont, UK) or enzymatically by Macrogen Inc. (Seoul, Korea). Sequencing reactions were performed in-house with an ABI Prism BIG Dye Terminator v.3.1 Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA, USA) or by either AGRF or Macrogen, followed by capillary separation by AGRF, Macrogen or the Applied Genetics Diagnostic Group (University of Melbourne, Australia).
Sequence editing and alignment
Contiguous sequences for each region were assembled and edited using Sequencher v.4.8 (Gene Codes Corporation, Ann Arbor, MI, USA). Edited sequences were aligned manually using Se-Al Sequence Alignment Editor v. 2.0a11 (Rambaut 2002). Gaps were inserted to ensure positional homology and to minimise nucleotide mismatches (Kelchner and Clark 1997). All variable nucleotide positions were checked against the original chromatogram files to ensure the accuracy of base calls.
Insertion–deletion events (INDELs) were coded using a method equivalent to the ‘simple coding’ of Simmons and Ochoterena (2000), with a single character representing each INDEL whether single- or multi-base. There was a deletion of 69 bp in the trnQ–5′rps16 intergenic spacer of two accessions of G. aquifolium from Roses Gap in the Grampians Ranges, Victoria. Because the sequence data corresponding to this deletion in other accessions contained several base substitutions and single-base insertions, all variable base positions were included, but with the two Roses Gap accessions scored as ‘missing’ for those positions. Also, in the trnQ–5′rps16 spacer, there is a short inverted (and complimented) region (Bases 346–350 in the alignment). Although initially included as a binary character, on closer examination, this region was found to be the loop of a putative stem–loop structure and homoplasious, and so was excluded from the final analyses. Alignments included four variable poly-N regions, two each in the trnL–trnF and rpoB–trnC spacers. Two were highly variable and homoplasious and were excluded from analysis. The other two were coded as multistate characters for phylogenetic analysis. Alignments used for analysis are available on request.
In an initial study, one accession of G. bedggoodiana (MELU D104178; not listed in Table 1) was found to contain a small number of polymorphic nucleotide sites distributed across the trnL–trnF and rpoB–trnC spacers. Heteroplasmy was confirmed by the re-isolation of DNA and subsequent sequencing of cloned copies of an rpoB–trnC segment (Holmes 2008). Data from this sample were not used in subsequent analyses.
Comparison of ITS paralogues
For several taxa in our dataset, we were able to amplify and sequence paralogous copies of ITS. Paralogues within and among individuals were compared, with a view to distinguishing potentially functional copies from pseudogenes. ITS-1 was highly divergent among paralogues and difficult to align; therefore, our comparisons focussed on the 5.8S gene and ITS-2. Comparisons included G/C content, the number of CpG and CpNpG methylation sites and the presence of conserved motifs, all of which have been shown to vary between functional and pseudogenic rDNA (Buckler et al. 1997; Bayly and Ladiges 2007). Sequences were examined for the presence of four motifs in the 5.8S gene previously identified as highly conserved in flowering-plant rDNA. These were as follows: 5′-GAATTGCAGAATC (Jobes and Thien 1997); an EcoRV restriction site (GATATC) near the 5′ end (Liston et al. 1996); the motifs M1 and M3 identified by Harpke and Peterson (2008). We also compared, among paralogues, the distribution of variable sites across the 5.8S gene and ITS-2. Boundaries between these regions were identified by comparisons with previously published sequences, including those for Macadamia (Proteaceae; Mast et al. 2008).
Phylogenetic analyses
Parsimony analyses were performed using PAUP* 4.0b10 (Swofford 2001). For all analyses, individual nucleotide sites (base positions) were coded as equally weighted, unordered multistate characters; missing data were coded as ‘?’, and gaps (coded in the matrix as ‘–’) were treated as a new state. Accessions exhibiting multiple states for a character (i.e. ambiguities) were interpreted as polymorphic at that character or position. Starting trees were obtained by stepwise addition, using a closest addition sequence (holding one tree at each step), and then subjected to tree-bisection–reconnection (TBR) branch-swapping, with the MULTREES option on, with no limit on the maximum number of trees to be saved (MaxTrees). All other options were left on their default settings. A strict consensus tree for each dataset was derived from the set of equally most parsimonious trees. Branch lengths were calculated for one of the equally most parsimonious trees using DELTRAN character state optimisation. The consistency index (CI) (Kluge and Farris 1969) and the retention index (RI) (Farris 1989) were calculated to determine the amount of homoplasy and synapomorphies within the cladogram (Kitching et al. 1998). Support for nodes was tested by bootstrap analysis, using a full heuristic search, with 1000 bootstrap replicates, starting by random stepwise addition, with TBR branch-swapping and saving no more than 5000 trees per addition replicate.
For the ITS regions, two separate datasets were analysed; the first compared divergent copies from a limited number of samples (putative functional copies and pseudogenes) and the second included only putatively functional copies across a broader sample of taxa. Outgroups were Grevillea papuana and G. robusta for the first analysis, and Hakea nodosa and H. ulicina for the second analysis.
Datasets from each cpDNA region were analysed separately and the results compared. Outgroups for all cpDNA analyses were G. acanthifolia, G. laurifolia and G. willisii. The incongruence length difference test (ILD; Farris et al. 1994, 1995) was used to test for congruence between phylogenetic signals across the different cpDNA regions. The ILD tests were implemented using the ‘partition homogeneity’ (HomPart) option in PAUP*, using only informative characters of each dataset. Tests were run using 1000 randomisations (homogeneity replicates) and a heuristic search to obtain the sum of tree lengths for data from each DNA region (Swofford 2001). The heuristic search options employed were start by stepwise addition, closest addition sequence, one tree held at each step and TBR branch-swapping. All other settings were left on default. All ILD tests were conducted on the University of Oslo Bioportal (Kumar et al. 2009), previously available at www.bioportal.uio.no (accessed 2012).
Results
ITS variation and phylogeny
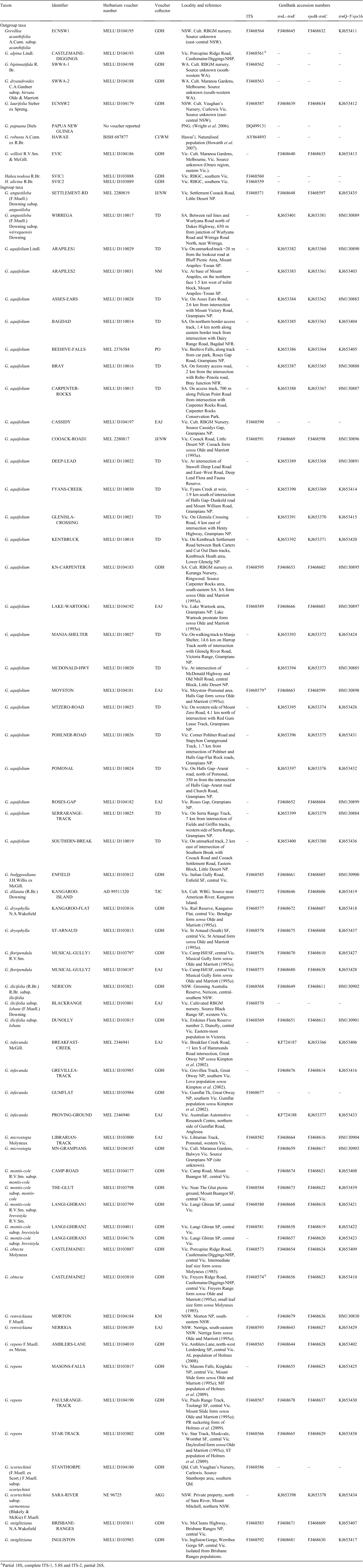
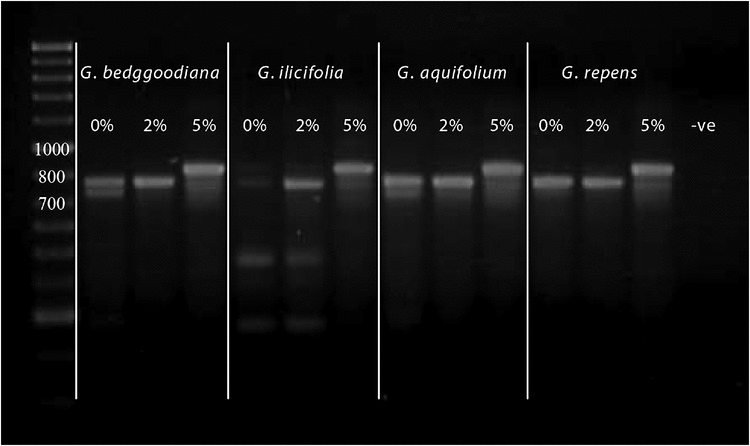
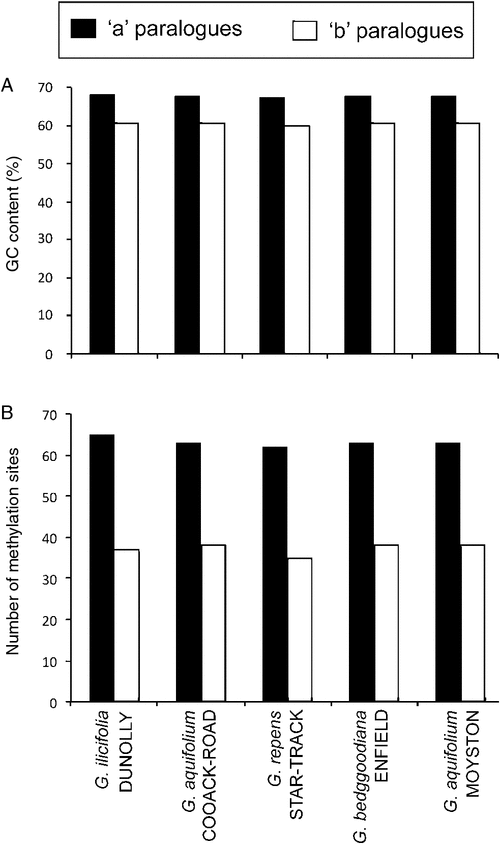
For many samples in our dataset, PCR amplification of the combined ITS region (including ITS-1, 5.8S, ITS-2) identified multiple fragment-length variants within individuals. The number and size of amplicons for any individual varied with PCR conditions, particularly with differing concentrations of dimethyl sulfoxide (DMSO) in the reactions (e.g. Fig. 2). Preferential amplification of the largest ‘a’ copy of this region was generally most efficient with the addition of 5–6% (v/v) DMSO, and the shorter ‘b’ copy with 1–2% (v/v) DMSO. In some cases, this approach allowed sequencing of the predominant ITS paralogue size classes without the need for cloning or gel extraction and was used to compare the 5.8S/ITS-2 regions for two of the largest distinct band sizes (‘a’ and ‘b’) for five ingroup accessions. The size classes fell into two divergent clades in the first phylogenetic analysis (Fig. 3). Characters separating the two clades were distributed across the 5.8S gene and ITS-2 (Fig. 4). Variation among ‘a’ copy sequences was low and restricted to sites in ITS-2, whereas variation among the shorter ‘b’ copies was greater and variable sites were distributed throughout the 5.8S and ITS-2 (Fig. 4). The 5.8S–ITS-2 portion of the sequence for the larger ‘a’ copy in each sample had a higher G/C content and higher number of methylation sites (Fig. 5). The ‘b’ copies also lacked the conserved angiosperm motif M1 in the 5.8S identified by Harpke and Peterson (2008). These features of the ‘b’ copies are characteristic of rDNA pseudogenes, as observed in many other plant groups (e.g. Buckler et al. 1997; Mayol and Roselló 2001; Bayly et al. 2008; Burke et al. 2008). Given this, our subsequent analyses focused purely on the ‘a’ copy amplified from a broader range of samples.
|

|
|
Parsimony analysis of the larger dataset of ‘a’ copy sequences yielded 5763 equally most parsimonious trees (length 42 steps, CI = 0.91, RI = 0.93). The holly grevilleas (Fig. 6) are supported as monophyletic and distinct from the outgroup and Grevillea alpina (in the Floribunda group of Makinson 2000). Branch lengths within the holly grevilleas were short and the strict consensus tree showed little resolution of relationships (Fig. 6), with all holly grevilleas being united by a large polytomous node. Within this polytomy, only three further nodes were identified with bootstrap support (BS) of >50%. One of these united the three samples of G. repens, one united the two samples of G. obtecta, and the third united the single accessions of G. bipinnatifida and G. dryandroides subsp. hirsuta.
cpDNA phylogeny
A complete dataset of sequences for the three cpDNA intergenic spacers studied (trnL–trnF, trnQ–5′rps16 and partial rpoB–trnC) was obtained for 57 samples (Table 1). The observed sequence characteristics for each of these regions are listed in Table 2. Results from the three DNA regions were highly congruent and the ILD tests were non-significant (P > 0.05) for all comparisons. Given this, and the fact that all regions are part of the same non-recombining chloroplast genome, only the results of a combined analysis of chloroplast regions are presented here.

|
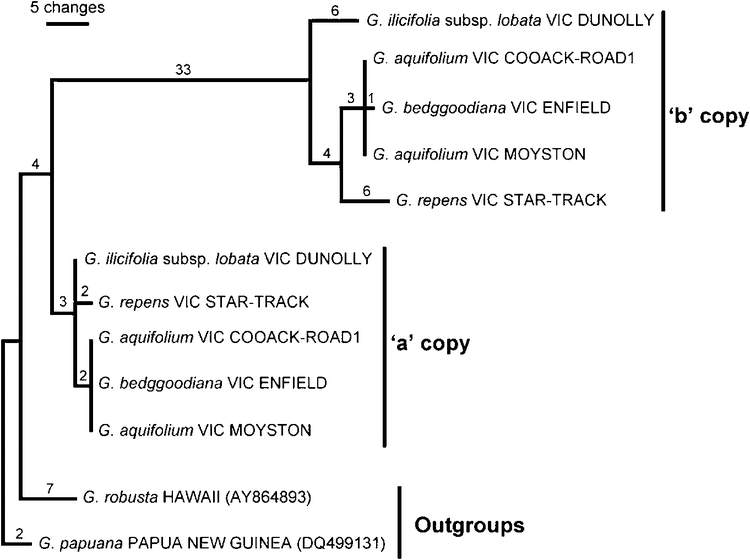
Parsimony analyses of the combined dataset recovered 60 equally most-parsimonious trees, with a length of 114, CI of 0.92 (CI excluding uninformative characters = 0.90) and RI of 0.97. One of the shortest trees, showing branch lengths and support values, is shown in Fig. 7. Only two of the nodes on this tree (indicated) were not present on the strict consensus; all others have BS of >50%.
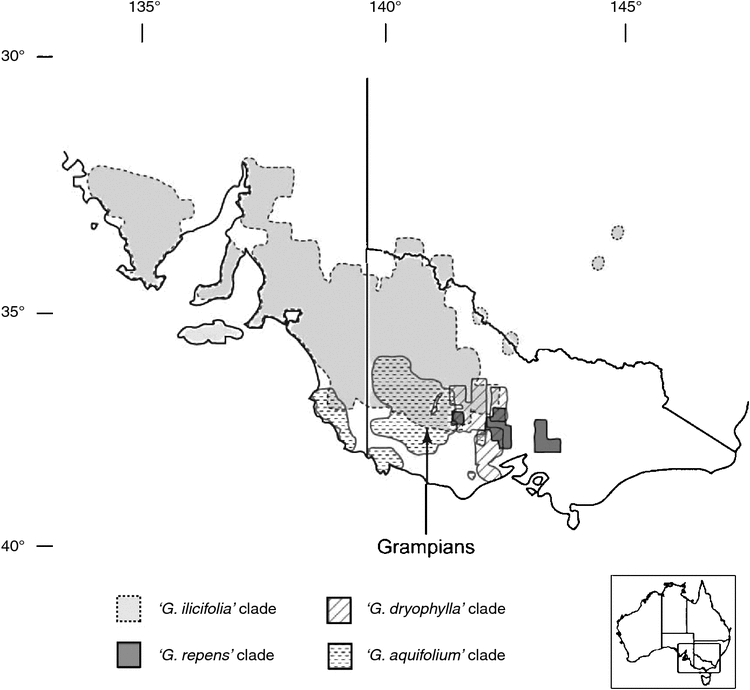
The ingroup taxa, together with one outgroup taxon, G. willisii, form a monophyletic group with 100% BS (Fig. 7, Node 1). G. willisii and the northern holly grevillea taxa, G. renwickiana (two accessions, Node 3) and G. scortechinii subsp. sarmentosa, are separate lineages that are unresolved in the polytomy at Node 1, whereas the other members of the ingroup form a monophyletic group with 100% BS (Fig. 7, Node 4). These are the southern holly grevilleas, which include the following four main clades: the ‘G. ilicifolia’, ‘G. dryophylla’, ‘G. repens’ and ‘G. aquifolium’. The geographic distributions of these clades are shown in Fig. 8. The ‘G. ilicifolia group’ includes G. ilicifolia, G. angustiloba and G. dilatata, and the clade has a strong BS (100%, Node 5). This clade, which has the broadest distribution and extends into South Australia, is the sister group to the remaining three clades of southern holly grevilleas (Node 7, with 94% BS). Of the three other clades, the ‘G. repens’ and ‘G. dryophylla’ clades of central Victoria have moderate (63%, Node 8) and high (92%, Node 13) BS respectively. The ‘G. aquifolium’ clade occurs west of these two groups and has moderate BS (63%, Node 18); however, the species G. aquifolium is not resolved as monophyletic (see Nodes 18 and 13).
|
The ‘G. repens’ clade (Node 8, Fig. 7) is composed of three taxa, namely, G. repens, G. obtecta and G. montis-cole subsp. brevistyla. Each taxon was confirmed as monophyletic, as follows: G. obtecta (Node 9, 78% BS), G. montis-cole subsp. brevistyla (Node 10, 91% BS) and G. repens (Node 11, 86% BS). G. repens (Nodes 11 and 12) exhibits some within-species differentiation, with two western Victoria accessions (locality codes AMBLERS-LANE and STAR-TRACK) being grouped at Node 12 with 86% BS; this is consistent with a genetic split between eastern and western populations in this species reported by Holmes et al. (2009) on the basis of nuclear microsatellite DNA data. G. montis-cole is not shown as monophyletic, with its two subspecies being placed in different clades (Nodes 10 and 15).
The ‘G. dryophylla’ clade (Node 13, Fig. 7) includes G. dryophylla, G. floripendula, G. steiglitziana, G. infecunda and G. montis-cole subsp. montis-cole, together with three accessions of G. aquifolium, each from a different location in the Grampians Ranges. Three characters, one from each of the intergenic spacers (trnL–trnF, trnQ–5′rps16, rpoB–trnC), support the placement of these three G. aquifolium accessions within the ‘G. dryophylla’ clade. All three characters are single-base substitutions and all have a CI = 1.0. Within the ‘G. dryophylla’ clade, the two accessions of G. montis-cole subsp. montis-cole cluster together (63% BS, Node 15). G. steiglitziana is resolved as non-monophyletic, with one accession clustering with G. dryophylla and G. floripendula at Node 17 (65% BS). The two accessions of G. steiglitziana are separated by two single-base substitutions (CI = 1.0), one from each of the rpoB–trnC and trnQ–5′rps16 spacers.
The ‘G. aquifolium’ clade (Node 18, 63% BS) includes the remaining G. aquifolium accessions in two main lineages on the strict consensus (Nodes 19 and 24), with G. microstegia being nested within one of them. Four accessions of G. aquifolium are unresolved in the polytomy at Node 18, where G. bedggoodiana is also positioned. These four G. aquifolium accessions are from different localities across the Grampians Ranges and areas immediately to the east. One lineage with strong support (Node 19, 93% BS) relates all G. aquifolium accessions from South Australia, the Little Desert and Mount Arapiles. There is evidence also of geographic differentiation in this clade, with the South Australian (Node 20) and Victorian (Node 21) accessions as sister clades. The second lineage (Node 24, 63% BS) includes seven accessions of G. aquifolium from central and western Grampians and south-western Victoria, with G. microstegia from Mount Cassell, eastern Grampians, nested within it (Node 27, 64% BS).
Discussion
This study has presented the first molecular phylogeny for any group of species in the genus Grevillea, one of Australia’s largest plant genera, and provided insight into the evolutionary relationships of the holly grevilleas. Circumscription of this group and delimitation of taxa has so far been based wholly on morphology, which shows complex patterns of variation. Molecular data here have identified major clades and their geographic distributions, allowing a comparison with current taxonomy and the distinctiveness of narrowly endemic species of conservation interest.
Monophyly of holly grevilleas
Without a phylogenetic framework for Grevillea, either morphological or molecular, identification of monophyletic groups is difficult. Our a priori circumscription of the holly grevilleas was based on the morphological groups of McGillivray and Makinson (1993) and Makinson (2000), and we chose three potentially related species, also from the Aspleniifolia–Hookeriana subgroup (Makinson 2000), as outgroups for the cpDNA analysis. Our cpDNA trees could not be rooted in such a way that the ingroup was monophyletic. Of the three outgroup species, two (G. acanthifolia and G. laurifolia) are more distinctive from the ingroup on the basis of both cpDNA sequences (longer branch lengths) and on morphology than is the third species, G. willisii. On this basis, we chose to root cpDNA trees on the branch connecting G. acanthifolia and G. laurifolia. Very low ITS sequence variation between these taxa and the ingroup species precluded their use as outgroups for the nrDNA analysis, for which two Hakea accessions were utilised.
Grevillea willisii, a species from eastern Victoria which is similar in cpDNA sequences to G. renwickiana and G. scortechinii (Fig. 7), exhibits overall morphology similar to that of the holly grevilleas, sharing characters such as pinnatipartite leaves (e.g. G. dryophylla, G. floripendula, G. obtecta, G. steiglitziana) and secund (toothbrush) conflorescences; the latter character is shared by all species included in the Pteridifolia group of Makinson (2000). However, G. willisii has a sessile ovary, differentiating it from the holly grevilleas, which all have a clearly stipitate, or stalked, ovary (McGillivray and Makinson 1993; Makinson 2000).
Exact relationships among the basal nodes of the cpDNA tree are unclear and G. willisii (outgroup) forms a polytomy with holly grevilleas on the strict consensus tree (Fig. 7). Resolving these relationships and testing the monophyly of the holly grevillea group would be best undertaken in the context of a broader phylogenetic analysis, using additional and more variable molecular markers. Including a wider sample of taxa from the Aspleniifolia–Hookeriana subgroup (Makinson 2000) in such a study would be worthwhile. In particular, G. pachylostyla, a segregate of, and presumed sister to, G. willisii (Olde and Marriott 1994; Makinson 2000) would be a valuable inclusion.
Major clades of holly grevilleas
The combined analysis of sequence data from three cpDNA regions provides insight into relationships of the holly grevilleas and, in particular, identifies several major clades. Of note is the strong support (the longest branch on the tree; 100% BS) for the southern holly grevilleas (Node 4, Fig. 7) forming a clade differentiated from G. willisii, G. renwickiana and G. scortechinii. This suggests an early geographic divergence of this clade (in central and western Victoria, South Australia and western New South Wales) from taxa occurring further east and north, mostly on and east of the Great Dividing Range (Fig. 1).
Among the southern holly grevilleas, the basal split, with strong support (Fig. 7), differentiates the G. ilicifolia clade from all other taxa. There has been some confusion between taxa of this group and G. aquifolium, including misidentification of specimens (Makinson 2000; Downing et al. 2004). Despite this, and despite some geographic overlap of the predominantly western G. ilicifolia clade with G. aquifolium and related species at the south-eastern portion of it range (Fig. 8), the cpDNA data support this clade being a distinct lineage and show no evidence of introgression or gene flow with G. aquifolium and close relatives. This is consistent with the morphological assessment of Downing et al. (2004).
In addition to the G. ilicifolia clade, the southern holly grevilleas include three closely related cpDNA clades, namely, the ‘G. repens’, ‘G. dryophylla’ and ‘G. aquifolium’ clades that range from central to western Victoria and south-eastern South Australia. These groups show some geographic differentiation (Fig. 8), which suggests the possibility of a vicariant history. What historical events this might relate to is a matter of speculation, but there is considerable climatic, topographic and edaphic variation across the region and a history of geological and climatic upheaval. For example, the Grampians Ranges (Fig. 8) were isolated by mid- and late-Miocene marine incursions into the lower Murray Basin, and more recent changes include Pleistocene climate fluctuations and substantial volcanic activity from the Pliocene to Holocene (e.g. Dodson 1974; Abele et al. 1976; Bowler et al. 1976; Costermans 1981; Nelson 1981; Marginson and Ladiges 1988; D’Costa et al. 1989; Crisp et al. 2001; Byrne 2008; Byrne et al. 2008; Pollock et al. 2013). Volcanic activity has potentially disrupted the distributions of holly grevilleas, which are effectively absent from volcanic soils. Likewise, aridity during Pleistocene glacials would have made some areas, especially north and west of the Grampians Ranges (Byrne 2008), less hospitable for holly grevilleas. Patterns of cpDNA variation in these plants might show evidence of vicariance events because the seeds have a limited capacity for dispersal, i.e. seeds are assumed to fall by gravity over short distances from parent plants, possibly with some secondary dispersal by ants for taxa such as G. repens that have elaiosomes.
cpDNA variation and species-level taxonomy
The combined analysis of three cpDNA regions did not resolve all species as monophyletic. This could be partly explained by a lack of divergence in the cpDNA regions sequenced (e.g. in the case of G. floripendula, G. dryophylla, G. infecunda, G. angustiloba and G. ilicifolia). However, there is clear incongruence between accepted species limits and the combined cpDNA tree (Fig. 7). Such patterns of incongruence are common in plants (e.g. McKinnon et al. 1999, 2001; Tsitrone et al. 2003; Meudt and Bayly 2008; Nevill et al. 2014), making the use of cpDNA sequences problematic for identification using a ‘DNA barcoding’ approach (e.g. Hollingsworth et al. 2011). Possible explanations for this incongruence include relatively low cpDNA mutation rates, homoplasy of DNA base changes, effects of incomplete chloroplast lineage sorting (e.g. the variable retention of ancestral cpDNA polymorphisms among descendant taxa), gene flow resulting from hybridisation–introgression (chloroplast capture) and poor taxonomy, the latter including excessive splitting, or reliance on homoplasious morphological traits.
For the holly grevilleas, it is unlikely that the mismatch between taxonomy and cpDNA variation is a result of genetic homoplasy (convergence of sequences). The potential for convergence in sequences among taxa arising by chance decreases as the number of character differences among lineages and number of DNA regions used increases (McKinnon et al. 1999, 2001). In our dataset, similar phylogenetic signal was observed in each of the three cpDNA regions, making genetic homoplasy an unlikely explanation for the observed patterns.
Incomplete chloroplast lineage sorting and/or hybridisation and introgression could affect some patterns of cpDNA variation observed in the holly grevilleas; distinguishing the influence of each of these processes is not straightforward, but evidence from the geographic distribution and relationships of cpDNA lineages can be used. If sharing of cpDNA haplotypes or lineages among species occurs in individuals from overlapping or proximal geographic areas, this is commonly taken as evidence of local introgression, as seen in eucalypts and other plant groups (e.g. Jackson et al. 1999; McKinnon et al. 1999, 2004; Pollock et al. 2013; Nevill et al. 2014); lineage sorting is not expected to produce such a geographic pattern (Schaal et al. 1998; Avise 2004). Gene flow associated with introgression can also result in the sharing of highly derived cpDNA lineages among species (e.g. among those located towards the tips of phylogenetic trees or haplotype networks), whereas incomplete lineage sorting is more likely to result in sharing of older, ancestral lineages (Schaal et al. 1998; Schaal and Leverich 2001).
Of particular interest here are the relationships and classification of four species, namely, G. microstegia, G. montis-cole, G. bedggoodiana and G. aquifolium. We will consider, in turn, what inferences can be made regarding these taxa.
Grevillea microstegia
Grevillea microstegia is confined to the top and slopes of Mount Cassell in the Mount William Range in the eastern Grampians (Molyneux 1975), where it grows over a few square kilometres. When first describing the species, Molyneux (1975) suggested that its closest morphological affinities were with G. floripendula (then undescribed) from the Ben Major area, near Beaufort, Victoria. However, in the cpDNA analysis G. microstegia is nested within the ‘G. aquifolium’ clade (Nodes 24–26; Fig. 7), clustered with G. aquifolium accessions from the Grampians and Lower Glenelg region in south-west Victoria. The closest G. aquifolium accession (Fyans Creek) to G. microstegia (Mount Cassell) differs from the latter by only two nucleotide substitutions. Specimens of G. microstegia can be distinguished from G. aquifolium by having more deeply lobed leaves, sometimes with tertiary lobing, and a sparser indumentum on the lower leaf surface (Makinson 2000; Downing 2012). However, Olde and Marriott (1995a) noted some minor variation in the size of leaf lobes in G. microstegia and reported the presence of putative hybrids of variable morphology, presumed to be between G. microstegia and G. aquifolium, on the lower slopes of Mount Cassell and nearby streams where the two species grow in close proximity.
The geographic range of G. microstegia is wholly enclosed within that of G. aquifolium and it is possible that G. microstegia is simply part of the spectrum of morphological variation within G. aquifolium, and that this is reflected in the observed pattern of cpDNA relationship. However, it is also possible that G. microstegia represents a separate lineage that has obtained a ‘G. aquifolium chloroplast’ by introgression; an effect of incomplete chloroplast lineage sorting seems less likely because of the geographic proximity of related haplotypes and their derived position in the phylogenetic tree (Fig. 7). Further sampling within G. microstegia, and of G. aquifolium populations on, and close to, Mount Cassell, and analysis using variable nuclear DNA markers would help confirm or refute the paraphyly of G. aquifolium inferred by our cpDNA data.
Grevillea montis-cole
Grevillea montis-cole was first described by Smith (1981), including two allopatric subspecies separated by ~10–15 km in western Victoria, namely, subsp. montis-cole from the Mount Buangor–Mount Cole area and subsp. brevistyla from neighbouring Mount Langi Ghiran. The two subspecies were not recognised by McGillivray and Makinson (1993), but were accepted by Makinson (1996, 2000), Olde and Marriott (1995b) and Walsh and Stasjic (2007). They are distinguished by characters such as style length (shorter in subsp. brevistyla) and leaf lobe and outline dimensions (Smith 1983; Olde and Marriott 1995b), and possibly occupy different niches; subsp. montis-cole is found as an understorey shrub in granitic loam soil, whereas subsp. brevistyla grows mainly in cracks and depressions in large granite outcrops at more exposed sites at higher altitude (Makinson 2000, G. D. Holmes, pers. obs.).
CpDNA data show distinction between the subspecies of G. montis-cole and other lineages sampled. All three cpDNA regions analysed show clear differences between the two subspecies (consistent with their taxonomic separation), but do not resolve the species as monophyletic (Fig. 7). Subsp. montis-cole is placed in the ‘G. dryophylla’ clade, which includes its suggested relatives on morphological grounds (G. steiglitziana and G. floripendula; Smith 1983), whereas subsp. brevistyla is placed in the ‘G. repens’ clade. This pattern of variation was unexpected for morphologically similar plants separated by just a few kilometres, and its basis is unclear. It could reflect historical hybridisation and introgression (e.g. between subsp. brevistyla and members of the ‘G. repens’ clade, or between subsp. montis-cole and the ‘G. dryophylla’ clade) or the stochastic effects of incomplete chloroplast lineage sorting. A population-level study using suitable nuclear markers would help elucidate the degree of differentiation between the two subspecies (e.g. whether they might warrant species-level recognition) and provide additional evidence of their relationships with other species.
Grevillea bedggoodiana
Grevillea bedggoodiana was first described by McGillivray (1986; attributed as J.H.Willis ex McGill.), and had been previously considered part of G. aquifolium, or an undescribed species allied to G. aquifolium or G. obtecta (Willis 1973; McGillivray and Makinson 1993). It is restricted to a small geographic range near Enfield, south of Ballarat, Victoria (Fig. 1), in an area with soil of marine sedimentary origin that has been encircled by newer volcanics. It is difficult to make firm inferences on the status of G. bedggoodiana because only one accession was included for analyses in the present study; however, in the combined cpDNA analysis it is related to G. aquifolium, being a genetically distinct lineage placed on the basal polytomy of the ‘G. aquifolium’ clade (Fig. 7). It is geographically disjunct from the main distribution of the ‘G. aquifolium’ clade and it may well be a distinct taxon whose history relates to allopatric peripheral isolation. However, greater population sampling and additional markers are required to confirm its phylogenetic position and degree of genetic differentiation from G. aquifolium. Additional study is also warranted to clarify the significance of the heteroplasmy identified in a second sample of G. bedggoodiana (Holmes 2008). Divergent cpDNA haplotypes may occur among (and within) individuals or populations of this species, which could affect inference of its phylogenetic placement.
Grevillea aquifolium
Grevillea aquifolium is a widespread (Fig. 1) and morphologically variable species (Olde and Marriott 1995a) and was a particular focus of sampling in the present study. Analysis of cpDNA placed the majority of samples in the ‘G. aquifolium’ clade, along with G. microstegia and G. bedggoodiana, as discussed above. However, three accessions of G. aquifolium were not resolved within this clade, but instead clustered within the ‘G. dryophylla’ clade (Node 13; Fig. 7). Three characters, one from each of three cpDNA regions (trnL–trnF, trnQ–5′rps16, rpoB–trnC), support the inclusion of these accessions in the ‘G. dryophylla’ clade. These accessions of G. aquifolium are each from different populations in the Grampians in western Victoria, an area where the species is very common. They are not morphologically distinct from surrounding plants or populations of G. aquifolium and they are geographically disjunct from other members of the ‘G. dryophylla’ clade, which occur in central Victoria (Fig. 8). This geographic pattern is not consistent with recent introgression between taxa in different clades, especially maternal (seed-mediated) gene flow between disjunct areas, but may result from historical introgression events between lineages from distinct clades if their ranges previously overlapped. Another plausible explanation for this pattern is related to incomplete chloroplast lineage sorting, i.e. that the distinct chloroplast clades predate the differentiation of at least some taxa and that representatives of both clades have been variably retained in different populations/individuals of G. aquifolium. If this is the case for G. aquifolium, it also means that such explanations might apply to other taxa of holly grevilleas that share related chloroplast haplotypes. Again, further data would help better understand the evolutionary history of the group, including nuclear markers and additional individuals/populations for taxa other than G. aquifolium.
Utility of ITS sequences for phylogeny reconstruction
Individual plant genomes include many copies of the ITS regions of nrDNA. These are commonly homogenised through concerted evolution (Arnheim 1983), such that there is little within-individual variation, allowing these DNA regions to be widely used in phylogenetic studies of flowering plants (Hershkovitz et al. 1999). However, numerous studies have reported the presence of divergent ITS paralogues within genomes, often including putative non-functional ‘pseudogenes’ (e.g. Buckler et al. 1997; Mayol and Roselló 2001; Bayly et al. 2008; Burke et al. 2008). This also appears to be the case in Grevillea. Presence of paralogues, when not recognised, can confound phylogeny reconstruction and lead to misleading phylogenetic trees (Sanderson and Doyle 1992; Bailey et al. 2003).
In the present study, we identified multiple ITS paralogues from PCR fragment-size and sequence comparisons. We attempted to compare only orthologues for phylogeny reconstruction, in particular using the presumed functional copies, based on sequence characteristics (G/C content, distribution of variable sites, number of methylation sites), as has been done in studies of other groups (e.g. Buckler et al. 1997; Bayly et al. 2008). Among the presumed functional orthologues, we found some phylogenetic signal, but there was inadequate resolution to separate closely related Grevillea lineages (Fig. 6), even those from different taxonomic subgroups (e.g. G. bipinnatifida v. G. aquifolium). Although the percentage of parsimony-informative sites relative to sequence length in the ITS was similar to that of cpDNA regions, much of this variation was found at nucleotide sites that were polymorphic within individuals. This level of within-genome variation makes the ITS regions highly problematic for use in phylogeny reconstruction in Grevillea, and most likely in other genera of Proteaceae (Mast and Givnish 2002).
Implications for conservation
All species of Grevillea in Victoria are protected under the Flora and Fauna Guarantee (FFG) Act; however, active conservation is dependent on priority listings. Our findings raise questions about the evolutionary distinctness of some recognised taxa and suggest a review of their conservation status. The differing level of DNA sequence variation observed among species of the G. aquifolium group hints at unrecognised taxa and possible instances of over-splitting. In particular, the large genetic differentiation between the two subspecies of G. montis-cole suggests that these lineages may constitute separate species. Pending a detailed population-genetic study, both lineages warrant recognition of conservation status that is currently afforded only to subsp. brevistyla. Considerable cpDNA and morphological variation exists within G. aquifolium, indicative of a long history of population isolation. Although there is evidence of cpDNA sequence divergence among lineages within G. aquifolium, we suggest that the current taxonomic status of this species should be maintained pending further examination. The species G. microstegia should also be examined in more detail to ascertain its taxonomic status because its highly restricted distribution defines it as an endangered species under the FFG Act (1988) and the EPBC Act (1999).
Acknowledgements
For assistance with field work or provision of specimens, we thank Greg Downing, Val Downing, Andrew Downing, Adele Gibbs, Pauline Ladiges, Neil Marriot, Laura Pollock, Adrian Holmes, Trevor Christensen, Peter Olde and Keith McDougall. Pauline Ladiges, Yvonne Parsons and Andrew Drinnan provided valuable advice during the PhD candidatures of TLD and GDH. Plant-collecting permits were provided by the Victorian Department of Sustainability and Environment, NSW National Parks and Wildlife Service and South Australian Department of Environment and Heritage and Forestry SA. This work was supported by the Hansjörg Eichler Scientific Research Fund award from the Australasian Systematic Botany Society (grant to TLD), the Holsworth Wildlife Research Endowment (grant to TLD) and the Cybec Foundation (funding to GDH). AAH thanks the Australian Research Council for fellowship support.
References
Abele C, Gloe CS, Hocking JB, Holdgate GR, Kenley PR, Lawrence CR, Ripper D, Threfall WF (1976) Chapter 8: Tertiary. In ‘Geology of Victoria’. (Eds JG Douglas, JA Ferguson) Special Publication Series of The Geological Society of Australia 5, pp. 177–274. (Geological Society of Australia Incorporated: Melbourne)Arnheim N (1983) Concerted evolution in multigene families. In ‘Evolution of Genes and Proteins’. (Eds M Nei, R Koehn) pp. 38–61. (Sinauer Associates: Sunderland, MA)
Avise JC (2004) ‘Molecular Markers, Natural History, and Evolution.’ (Sinauer Associates: Sunderland, MA)
Bailey CD, Carr TG, Harris SA, Hughes CE (2003) Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molecular Phylogenetics and Evolution 29, 435–455.
Bayly MJ, Ladiges PY (2007) Divergent paralogues of ribosomal DNA in eucalypts (Myrtaceae). Molecular Phylogenetics and Evolution 44, 346–356.
| Divergent paralogues of ribosomal DNA in eucalypts (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmtVOltbo%3D&md5=24cc2d7757e4ed6f840b7e97983ff1a6CAS | 17188000PubMed |
Bayly MJ, Udovicic F, Gibbs AK, Parra-O. C, Ladiges PY (2008) Ribosomal DNA pseudogenes are widespread in the eucalypt group (Myrtaceae): implications for phylogenetic analysis. Cladistics 24, 131–146.
| Ribosomal DNA pseudogenes are widespread in the eucalypt group (Myrtaceae): implications for phylogenetic analysis.Crossref | GoogleScholarGoogle Scholar |
Bentham G (1870) Grevillea. In ‘Flora Australiensis: A Description of the Plants of the Australian Territory. Vol. 5 (Myoporineae to Proteaceae)’. pp. 417–489. (L. Reeve & Co.: London)
Bowler JM, Hope GS, Jennings JN, Singh G, Walker D (1976) Late Quaternary climates of Australia and New Guinea. Quaternary Research 6, 359–394.
| Late Quaternary climates of Australia and New Guinea.Crossref | GoogleScholarGoogle Scholar |
Briggs JD, Leigh JH (1996) ‘Rare or threatened Australian plants.’ (CSIRO Publishing: Melbourne)
Buckler ES, Ippolito A, Holtsford TP (1997) The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics 145, 821–832.
Burke JM, Bayly MJ, Adams P, Ladiges PY (2008) Molecular phylogenetic analysis of Dendrobium (Orchidaceae), with emphasis on the Australian section Dendrocoryne, and implications for generic classification. Australian Systematic Botany 21, 1–14.
| Molecular phylogenetic analysis of Dendrobium (Orchidaceae), with emphasis on the Australian section Dendrocoryne, and implications for generic classification.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXktFyit70%3D&md5=04e5159e034585931593dc51e9056dbeCAS |
Byrne M (2008) Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography. Quaternary Science Reviews 27, 2576–2585.
| Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Yeates DK, Joseph L, Kearney M, Bowler J, Williams MA, Cooper S, Donnellan SC, Keogh JS, Leys R, Melville J, Murphy DJ, Porch N, Wyrwoll KH (2008) Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota. Molecular Ecology 17, 4398–4417.
| Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1cjhvFGruw%3D%3D&md5=1e310eccbf8e4218d3a53861be7501f9CAS | 18761619PubMed |
Costermans LF (1981) ‘Native Trees and Shrubs of South-eastern Australia.’ (Rigby Publishers: Sydney)
Crisp MD, Laffan S, Linder HP, Monro A (2001) Endemism in the Australian flora. Journal of Biogeography 28, 183–198.
| Endemism in the Australian flora.Crossref | GoogleScholarGoogle Scholar |
D’Costa DM, Edney P, Kershaw AP, De Deckker P (1989) Late Quaternary palaeoecology of Tower Hill, Victoria, Australia. Journal of Biogeography 16, 461–482.
| Late Quaternary palaeoecology of Tower Hill, Victoria, Australia.Crossref | GoogleScholarGoogle Scholar |
Dodson JR (1974) Vegetation history and water fluctuations at Lake Leake, south-eastern South Australia. I. 10 000 B.P. to present. Australian Journal of Botany 22, 719–741.
| Vegetation history and water fluctuations at Lake Leake, south-eastern South Australia. I. 10 000 B.P. to present.Crossref | GoogleScholarGoogle Scholar |
Downing TL (2012) Investigating genetic and morphological variability in the holly grevillea Grevillea aquifolium (Proteaceae). PhD Thesis, School of Botany, The University of Melbourne.
Downing TL, Duretto MF, Ladiges PY (2004) Morphological analysis of the Grevillea ilicifolia complex (Proteaceae) and recognition of taxa. Australian Systematic Botany 17, 327–341.
| Morphological analysis of the Grevillea ilicifolia complex (Proteaceae) and recognition of taxa.Crossref | GoogleScholarGoogle Scholar |
Farris JS (1989) The retention index and homoplasy excess. Systematic Zoology 38, 406–407.
| The retention index and homoplasy excess.Crossref | GoogleScholarGoogle Scholar |
Farris JS, Källersjö M, Kluge AG, Bult C (1994) Testing significance of incongruence. Cladistics 10, 315–319.
| Testing significance of incongruence.Crossref | GoogleScholarGoogle Scholar |
Farris JS, Källersjö M, Kluge AG, Bult C (1995) Constructing a significance test for incongruence. Systematic Biology 44, 570–572.
| Constructing a significance test for incongruence.Crossref | GoogleScholarGoogle Scholar |
George AS (1998) Proteus in Australia. An overview of the current state of taxonomy of the Australian Proteaceae. Australian Systematic Botany 11, 257–266.
| Proteus in Australia. An overview of the current state of taxonomy of the Australian Proteaceae.Crossref | GoogleScholarGoogle Scholar |
Harpke D, Peterson A (2008) 5.8S motifs for the identification of pseudogenic ITS regions. Botany 86, 300–305.
| 5.8S motifs for the identification of pseudogenic ITS regions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXltVKhs7k%3D&md5=0c81429af03ad4484ca7dcbb13b09c83CAS |
Hershkovitz MA, Zimmer EA, Hahn WJ (1999) Ribosomal DNA sequences and angiosperm systematics. In ‘Molecular Systematics and Plant Evolution’. (Eds PM Hollingsworth, RM Bateman, RJ Gornall) pp. 268–326. (Taylor and Francis: London)
Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS ONE 6, e19254
| Choosing and using a plant DNA barcode.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXntVajt7Y%3D&md5=986cbaec3cad31cdc9cc9daf5688d4ebCAS | 21637336PubMed |
Holmes GD (2008) Conservation biology of the rare shrub Grevillea repens F.Muell. ex Meisn. (Proteaceae). PhD Thesis, Centre for Environmental Stress and Adaption Research (CESAR; Department of Genetics) and School of Botany, The University of Melbourne.
Holmes GD, James EA, Hoffmann AA (2009) Divergent levels of genetic variation and ploidy among populations of the rare shrub, Grevillea repens (Proteaceae). Conservation Genetics 10, 827–837.
| Divergent levels of genetic variation and ploidy among populations of the rare shrub, Grevillea repens (Proteaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXntlShu78%3D&md5=89d68098a2ffd647f849a2532f23c023CAS |
Howarth FG, James SA, McDowell W, Preston DJ, Imada CT (2007) Identification of roots in lava tube caves using molecular techniques: implications for conservation of cave arthropod faunas. Journal of Insect Conservation 11, 251–261.
| Identification of roots in lava tube caves using molecular techniques: implications for conservation of cave arthropod faunas.Crossref | GoogleScholarGoogle Scholar |
Jackson HD, Steane DA, Potts BM, Vaillancourt RE (1999) Chloroplast DNA evidence for reticulate evolution in Eucalyptus (Myrtaceae). Molecular Ecology 8, 739–751.
James EA (2000) Implications of reproductive biology and morphology on the conservation of Grevillea williamsonii (Proteaceae). Final report to Australian Flora Foundation. Available at http://www.aff.org.au/James_Gwilliamsonii_final.pdf [Verified 22 May 2014]
James EA (2004) Conserving Grevillea williamsonii: the importance of taxonomic research for appropriate conservation action. BGjournal 1, 17–19.
James EA, McDougall KL (2014) Spatial genetic structure reflects extensive clonality, low genetic diversity and habitat fragmentation in Grevillea renwickiana (Proteaceae), a rare, sterile shrub from south-eastern Australia. Annals of Botany
| Spatial genetic structure reflects extensive clonality, low genetic diversity and habitat fragmentation in Grevillea renwickiana (Proteaceae), a rare, sterile shrub from south-eastern Australia.Crossref | GoogleScholarGoogle Scholar | 24737718PubMed |
Jobes DV, Thien LB (1997) A conserved motif in the 5.8S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences. Plant Molecular Biology Reporter 15, 326–334.
| A conserved motif in the 5.8S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXjs1arurY%3D&md5=dbe02ec81129b1ed4cd94f1fe6346db4CAS |
Kelchner SA, Clark LG (1997) Molecular evolution and phylogenetic utility of the chloroplast rpl16 intron in Chusquea and the Bambusoideae (Poaceae). Molecular Phylogenetics and Evolution 8, 385–397.
| Molecular evolution and phylogenetic utility of the chloroplast rpl16 intron in Chusquea and the Bambusoideae (Poaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXjtVensw%3D%3D&md5=f9edcc76a21f2a57328ed8f27317fa75CAS | 9417896PubMed |
Kimpton SK, James EA, Drinnan AN (2002) Reproductive biology and genetic marker diversity in Grevillea infecunda (Proteaceae), a rare plant with no known seed production. Australian Systematic Botany 15, 485–492.
| Reproductive biology and genetic marker diversity in Grevillea infecunda (Proteaceae), a rare plant with no known seed production.Crossref | GoogleScholarGoogle Scholar |
Kitching IJ, Forey PL, Humphries CJ, Williams DM (1998) ‘Cladistics: The Theory and Practice of Parsimony Analysis.’ (Oxford University Press: Oxford, UK)
Kluge AG, Farris JS (1969) Quantitative phyletics and the evolution on Anurans. Systematic Zoology 18, 1–32.
| Quantitative phyletics and the evolution on Anurans.Crossref | GoogleScholarGoogle Scholar |
Kumar S, Skjæveland Å, Orr RJS, Enger P, Ruden T, Mevik B-J, Burki F, Botnen A, Shalchian-Tabrizi K (2009) AIR: a batch-oriented web program package for construction of super matrices ready for phylogenetic analyses. BMC Bioinformatics 10, 357
| AIR: a batch-oriented web program package for construction of super matrices ready for phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar | 19863793PubMed |
Liston A, Robinson WA, Oliphant JM, Alvarez-Buylla ER (1996) Length variation in the nuclear ribosomal DNA internal transcribed spacer region of non-flowering seed plants. Systematic Botany 21, 109–120.
| Length variation in the nuclear ribosomal DNA internal transcribed spacer region of non-flowering seed plants.Crossref | GoogleScholarGoogle Scholar |
Makinson RO (1996) Grevillea. In ‘Flora of Victoria. Vol. 3’. (Eds NG Walsh, TJ Entwisle) pp. 845–870. (Inkata Press: Melbourne)
Makinson RO (2000) Grevillea. In ‘Flora of Australia. Vol. 17A: Proteaceae 2’. (Ed. AJG Wilson) pp. 1–460. (CSIRO Publishing: Melbourne)
Marginson JC, Ladiges PY (1988) Geographical variation in Eucalyptus baxteri s.l. and the recognition of a new species, E. arenacea. Australian Systematic Botany 1, 151–170.
Mast MR, Givnish TJ (2002) Historical biogeography and the origin of stomatal distributions in Banksia and Dryandra (Proteaceae) based on their cpDNA phylogeny. American Journal of Botany 89, 1311–1323.
| Historical biogeography and the origin of stomatal distributions in Banksia and Dryandra (Proteaceae) based on their cpDNA phylogeny.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXislejsw%3D%3D&md5=978238a1894d5bba0e1f498ba5a37219CAS |
Mast AR, Willis CL, Jones EH, Downs KM, Weston PH (2008) A smaller Macadamia from a more vagile tribe: inference of phylogenetic relationships, divergence times, and diaspore evolution in Macadamia and relatives (tribe Macadamieae; Proteaceae). American Journal of Botany 95, 843–870.
| A smaller Macadamia from a more vagile tribe: inference of phylogenetic relationships, divergence times, and diaspore evolution in Macadamia and relatives (tribe Macadamieae; Proteaceae).Crossref | GoogleScholarGoogle Scholar | 21632410PubMed |
Mayol M, Roselló JA (2001) Why nuclear ribosomal spacers (ITS) tell different stories in Quercus. Molecular Phylogenetics and Evolution 19, 167–176.
| Why nuclear ribosomal spacers (ITS) tell different stories in Quercus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXjt1Gmsb8%3D&md5=5265945f2c243d3d6c970360587389f1CAS | 11341800PubMed |
McGillivray DJ (1986) ‘New Names in Grevillea (Proteaceae).’ (Published and distributed by the author: Sydney) [Facsimile in McGillivray and Makinson (1993)]
McGillivray DJ, Makinson RO (1993) ‘Grevillea, A Taxonomic Revision.’ (Melbourne University Press: Melbourne)
McKinnon GE, Steane DA, Potts BM, Vaillancourt RE (1999) Incongruence between chloroplast and species phylogenies in Eucalyptus subgenus Monocalyptus (Myrtaceae). American Journal of Botany 86, 1038–1046.
McKinnon GE, Vaillancourt RE, Jackson HD, Potts BM (2001) Chloroplast sharing in the Tasmanian eucalypts. Evolution 55, 703–711.
McKinnon GE, Jordan GJ, Vaillancourt RE, Steane DA, Potts BM (2004) Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359, 275–284.
Meudt HM, Bayly MJ (2008) Phylogeographic patterns in the Australasian genus Chionohebe (Veronica s.l., Plantaginaceae) based on AFLP and chloroplast DNA sequences. Molecular Phylogenetics and Evolution 47, 319–338.
| Phylogeographic patterns in the Australasian genus Chionohebe (Veronica s.l., Plantaginaceae) based on AFLP and chloroplast DNA sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXktV2jtrc%3D&md5=272f271a0ee6137ba055560283b7b2cdCAS | 18299210PubMed |
Molyneux WM (1975) A new Grevillea species from western Victoria. Muelleria 3, 141–145.
Molyneux WM (1985) Grevillea obtecta (Proteaceae), a new species from central Victoria. Muelleria 6, 147–151.
Nelson EC (1981) Phytogeography of Southern Australia. In ‘Ecological Biogeography of Australia’, Vol. 1 (Ed. A Keast) pp. 734–759. (Dr W. Junk Publishers: The Hague, the Netherlands)
Nevill PG, Despres T, Bayly MJ, Bossinger G, Ades PK (2014) Shared phylogeographic patterns and widespread chloroplast haplotype sharing in Eucalyptus species with different ecological tolerances. Tree Genetics & Genomes
| Shared phylogeographic patterns and widespread chloroplast haplotype sharing in Eucalyptus species with different ecological tolerances.Crossref | GoogleScholarGoogle Scholar |
Olde P, Marriott N (1994) ‘The Grevillea book. Vol. 1.’ (Kangaroo Press: Sydney)
Olde P, Marriott N (1995a) ‘The Grevillea book. Vol. 2.’ (Kangaroo Press: Sydney)
Olde P, Marriott N (1995b) ‘The Grevillea book. Vol. 3.’ (Kangaroo Press: Sydney)
Pollock LJ, Bayly MJ, Nevill PG, Vesk PA (2013) Chloroplast DNA diversity associated with protected slopes and valleys for hybridizing Eucalyptus species on isolated ranges in south-eastern Australia. Journal of Biogeography 40, 155–167.
| Chloroplast DNA diversity associated with protected slopes and valleys for hybridizing Eucalyptus species on isolated ranges in south-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Rambaut A (2002) Se-Al: sequence alignment editor, version 2.0a11. Available at http://tree.bio.ed.ac.uk/software/ [Verified 22 May 2014]
Sanderson MJ, Doyle JJ (1992) Reconstruction of organismal and gene phylogenies from data on multigene families: concerted evolution, homoplasy, and confidence. Systematic Biology 41, 4–17.
| Reconstruction of organismal and gene phylogenies from data on multigene families: concerted evolution, homoplasy, and confidence.Crossref | GoogleScholarGoogle Scholar |
Sang T, Crawford DJ, Stuessy TF (1995) Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution. Proceedings of the National Academy of Sciences of the United States of America 92, 6813–6817.
| Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXntVSksLw%3D&md5=1e2ec5a6db5d436d151bcad2d35d7128CAS | 7624325PubMed |
Schaal BA, Leverich WJ (2001) Plant population biology and systematics. Taxon 50, 679–695.
| Plant population biology and systematics.Crossref | GoogleScholarGoogle Scholar |
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Molecular Ecology 7, 465–474.
| Phylogeographic studies in plants: problems and prospects.Crossref | GoogleScholarGoogle Scholar |
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany 92, 142–166.
| The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXht1Klsbc%3D&md5=b689305626d9fcff7aa95c9109209391CAS | 21652394PubMed |
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and hare III. American Journal of Botany 94, 275–288.
| Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and hare III.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXktFOjsLg%3D&md5=e2e5ab07b9c93276ab570f470d94df24CAS | 21636401PubMed |
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49, 369–381.
| Gaps as characters in sequence-based phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD38zntlKjtg%3D%3D&md5=28e47d46829cef3c7826a15c80c9a1c4CAS | 12118412PubMed |
Smith RV (1981) A new species of Grevillea (Proteaceae) from Victoria. Muelleria 4, 423–427.
Smith RV (1983) Grevillea montis-cole sp. nov. (Proteaceae) from Victoria. Muelleria 5, 223–227.
Swofford DL (2001) ‘PAUP*. Phylogenetic analysis using parsimony, version 4.0b10.’ (Illinois Natural History Survey, Smithsonian Institution: Champaign, IL)
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17, 1105–1109.
| Universal primers for amplification of three non-coding regions of chloroplast DNA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38Xhslel&md5=5ea10efc62b8cab58c7ae3beddbc4b57CAS | 1932684PubMed |
Tsitrone A, Kirkpatrick M, Levin DA (2003) A model for chloroplast capture. Evolution 57, 1776–1782.
Walsh NG, Stasjic V (2007) ‘A Census of the Vascular Plants of Victoria’, 8th edn. (Royal Botanic Gardens Melbourne: Melbourne)
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In ‘PCR Protocols: A Guide to Methods and Applications’. (Eds M Innis, D Gelfand, J Sninsky, T White) pp. 315–322. (Academic Press: San Diego, CA)
Willis JH (1973) Grevillea. In ‘A Handbook to Plants in Victoria. Vol. 2: Dicotyledons’. pp. 38–48. (Melbourne University Press: Melbourne)
Wright S, Keeling J, Gillman L (2006) The road from Santa Rosalia: a faster tempo of evolution in tropical climates. Proceedings of the National Academy of Sciences of the United States of America 103, 7718–7722.
| The road from Santa Rosalia: a faster tempo of evolution in tropical climates.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XlsVGksro%3D&md5=aa9d6642fe6b371637a282dcfb814e46CAS | 16672371PubMed |