An expanded phylogenetic analysis of Sannantha (Myrtaceae) and description of a new species
Peter G. Wilson A B and Margaret M. Heslewood AA National Herbarium of New South Wales, Royal Botanic Gardens and Domain Trust, Mrs Macquaries Road, Sydney NSW 2000, Australia.
B Corresponding author. Email: peter.wilson@rbgsyd.nsw.gov.au
Australian Systematic Botany 27(1) 78-84 https://doi.org/10.1071/SB14011
Submitted: 1 April 2014 Accepted: 26 May 2014 Published: 30 June 2014
Abstract
Sannantha is a genus of shrubs widely distributed in eastern Australia and New Caledonia. We added five taxa to a previously published molecular dataset, four from Australia and the fifth from New Caledonia, a total of 11 of the 16 species in the genus. One of the Australian taxa added is a new species apparently restricted to the Goonoowigall State Conservation Area near Inverell, New South Wales. The results of the molecular analysis are discussed in light of morphology and geographic distribution. The new species, Sannantha whitei Peter G.Wilson, is described.
Introduction
The genus Sannantha was established by Wilson (in Wilson et al. 2007) to accommodate several species included in Babingtonia by Bean (1997, 1999), which would have been referred to Baeckea or B. virgata sens. lat. in earlier publications (e.g. Wilson 1991). Sannantha is distinguished by having a cymose inflorescence, mostly with some axes reduced so as to appear umbel-like and, as a consequence, with bracteoles clustered at the apex of the peduncle. The previously published chloroplast and nuclear ETS phylogenies (Wilson et al. 2007) showed very strong support for the genus and also found good support (>80% bootstrap) for a close relationship between S. cunninghamii and S. crenulata, both of which have leaves with irregular margins. The analysis indicated moderate support (~75–80% bootstrap) for the relationship between the two New Caledonian taxa sampled, S. virgata sens. str. and S. leratii.
At the time the new generic name was published, the first author was aware of an undescribed species first collected in 1992 from the Goonoowigall State Conservation Area. However, specimens of this taxon lacked flowers and a full description could not be prepared. Recently, good fertile material was collected and this enabled detailed comparisons to be made with other named species, and also provided DNA material for an assessment of its relationships with other species.
Materials and methods
Molecular analysis
Five species of Sannantha, including the undescribed taxon, were added to an existing dataset featuring Sannantha and several closely related genera sampled from an earlier study re-evaluating eastern Australian ‘Babingtonia’ species (Table 1). In total, 11 of 16 Sannantha species were sampled. Taxa were sequenced for the following four chloroplast regions: the atpβ–rbcL intergenic spacer (S) and part of the trnK intron partitioned into the matK gene (M), and the contiguous adjacent 5′ (5M) and 3′ (3M) regions of the intron; and also for the external transcribed spacer (E) of the nuclear encoded RNA region. In addition, five sequences of the trnK intron region used in the previous analysis were extended for the present study. This included a corrected matK sequence for S. angusta after the discovery that an erroneous sequence for the 5′ trnK intron had been appended to the matK region for this taxon in our initial analysis. The updated sequence did not affect generic placement but improved resolution and support for this taxon within the Sannantha clade.

|
Methods for DNA extraction and amplification were those cited by Wilson et al. (2007), except that the forward primer myrtF, CTCCGTGCTGGTGCATCGAACTGC (Lucas et al. 2007), was used for most of the new ETS sequences. Sequences were run on an Applied Biosystems 3730 DNA Analyzer at the Ramaciotti Centre, University of New South Wales. Raw sequences were processed using ABI software Sequence Navigator v1.0.1 and aligned by eye in PAUP*4.0b10 (Swofford 2002). Insertions/deletions (indels) were positioned so as to best conform to the indel types of Golenberg et al. (1993) and, where informative, coded in MacClade version 4.08 (Maddison and Maddison 2000) according to the scheme of Simmons and Ochoterena (2000). Heuristic searches were conducted using PAUP*4.0b10 (Swofford 2002), using tree bisection reconnection branch-swapping to recover the most-parsimonious (MP) trees. Multiple replicates of random taxon addition searching were conducted, so as to detect multiple islands of trees, with subsequent use of the ‘condense’ option to delete duplicate trees. Multistate characters were treated as polymorphisms and swapping was on best trees. Supports were calculated using jackknife rather than bootstrap resampling, following the recommendations of Simmons and Freudenstein (2011). For jackknife analyses, 10 000 replicates of faststep searching were conducted, in which each replicate used random-taxon addition, no branch swapping, and the percentage of characters deleted was set at 33%.
The MP phylogenies generated were compared to those obtained using the Markov chain Monte Carlo (MCMC) method implemented in MrBayes 3.2 (Ronquist et al. 2012). The most appropriate substitution models to apply in likelihood-based analyses were determined using the AIC option in MrModelltest 2.3 (Nylander 2004), with data partitioned into the five regions indicated above and excluding the scored indels. The F81 substitution model was applied to region 3M, with the number of substitution types (nst) set to 1. The remaining four regions fit general time-reversible likelihood (GTR) models (nst = 6), with among-site rate variation equal (5M and S) or modelled by a gamma distribution (M and E), with four rate categories. Parameters were set to be unlinked and with rates variable between partitions, with all other priors for the analysis set flat (i.e. as Dirichlet priors). Two independent runs of 10 million generations using four chains were performed with tree sampling every 1000 generations. Trees generated before the four Markov chains reaching stationarity (the burn-in ~10%) were discarded. The remaining trees were used to construct a 50% majority-rule consensus tree in PAUP*, with posterior probabilities for nodes compared to those with jackknife support in the parsimony analyses.
Results
Phylogeny
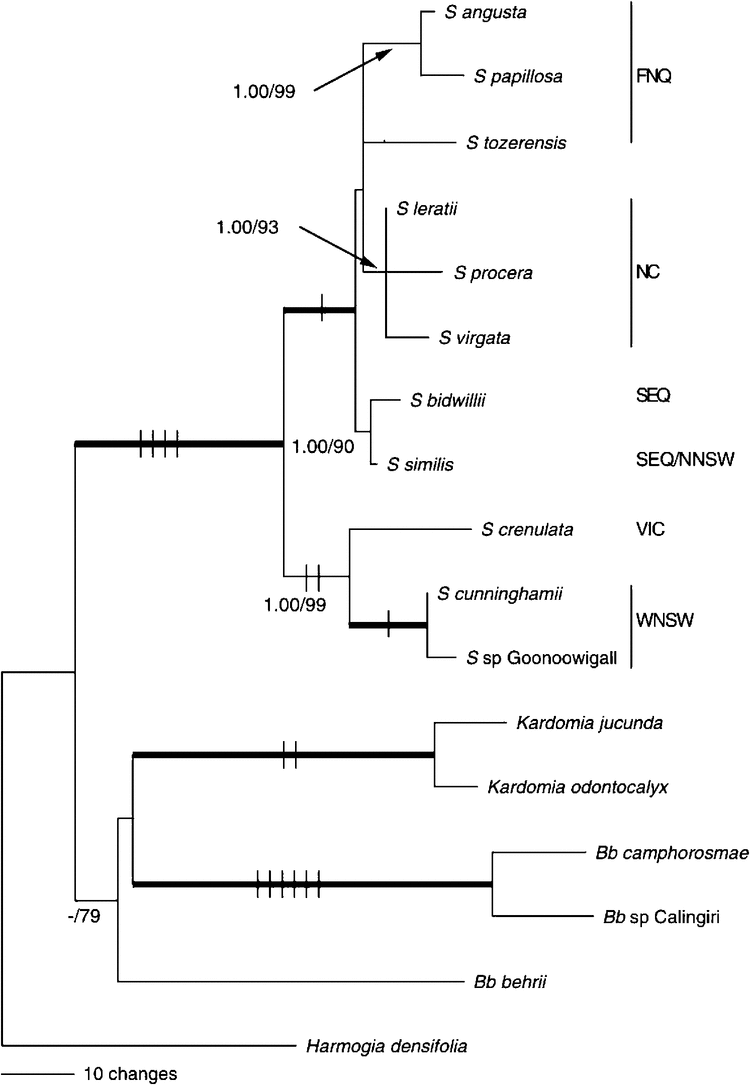
The heuristic search of the combined dataset yielded six equally most-parsimonious (MP) trees of 374 steps, with 170 parsimony-informative characters and 139 parsimony-uninformative characters from a total of 4017 characters. This included 18 scored indels, namely, 1 (5M), 1 (M), 4 (3M), 4 (S) and 8 (E); these were primarily single-base insertions or deletions, but four were repeats of adjacent sequence (2, 3, 5 and 12 bp in length), and one was a 4-bp inversion in the matK gene that was unique to Sannantha. A 4-bp deletion of a repeat unit (5M) shared by Sannantha and Kardomia is likely to have arisen independently.
The MrBayes consensus tree is shown in Fig. 1. The only structural difference from the MP strict consensus is the absence of a poorly supported sister relationship between Kardomia and Babingtonia behrii (69% jackknife). The analysis of the expanded dataset shows a robustly supported Sannantha (100% jk, 1.00 pp), with two strongly supported subclades. The clade is also defined by four indels, namely, two 1-bp insertions (E), a 7-bp deletion (S) and a 4-bp inversion (M)). The smaller of these two subclades was very strongly supported (99% jk, 1.00 pp, 1-bp insertion (E) and a 5-bp repeat (3M)) and comprises Sannantha crenulata, sister to a very strongly supported grouping of S. sp. Goonoowigall and S. cunninghamii (100% jk, 1.00 pp, 12-bp repeat (S)). The larger subclade, which is robustly supported but only partially resolved (100% jk, 1.00 pp, 1-bp insertion (E)), includes the other eight Sannantha species sampled, including the three New Caledonian and five Australian taxa. The two southern Queensland species, S. similis and S. bidwillii, form a well supported clade (90% jk, 1.00 pp), sister to the remaining six taxa that form an unsupported polytomy. Within the polytomy, the three New Caledonian species (93% jk, 1.00 pp) are strongly supported sisters, as are S. angusta and S. papillosa from far-northern Queensland (99% jk, 1.00 pp), with the position of S. tozerensis, also from far-northern Queensland, unresolved.
Separate analyses of data from the chloroplast and nuclear regions (data not shown) were consistent with the combined analysis, but with the nuclear-only data failing to resolve relationships within the larger Sannantha subclade.
Discussion
The topology of the Sannantha clade is similar to that found in the ETS analysis in our previous study (Wilson et al. 2007). The phylogenetic distance between the two subclades suggests that they diverged considerably earlier than the dispersal event that established the genus in New Caledonia. A fossil-dated chronogram of Myrtaceae by Thornhill et al. (2012) included a single representative of each of the two Sannantha subclades (‘Babingtonia’ cunninghamii and ‘Babingtonia’ tozerensis). The Thornhill study used both macro- and microfossils to date an analysis of an extensive sampling of the entire family. These authors used syncolpate pollen to estimate the date of the most recent common ancestor of the Leptospermeae+Chamelaucieae clades. This resulted in estimated ages with wide credibility intervals, so that the divergence of the two Sannantha species was dated as 11.82 million years ago, but with 95% credibility intervals of 3.98–20.74 million years ago (A. H. Thornhill, pers. comm.), placing the branching event in the Miocene. Given that calibration of divergence times within Sannantha would be reliant on the derived Thornhill date, we did not see that conducting an independent dating analysis would produce more precise age estimates on internal branches.
Species in the larger subclade, including S. tozerensis from the Iron Range in Cape York, are predominantly found in north-eastern Australia in coastal or tableland areas, whereas species in the smaller subclade, including S. cunninghamii from the drier western slopes of New South Wales, occur well inland in south-eastern Australia, but in areas with locally wetter habitats within the landscape. So the divergence of the two lineages represented by these taxa correlates reasonably well with evidence suggesting that aridity in inland Australia began to develop from the mid- to late Miocene (Martin 2006). Within the smaller subclade, S. cunninghamii has the most extensive range (~400 km), whereas the other two species apparently occur in more mesic refugia, with S. crenulata occurring to the south of this range in the Victorian alpine region and S. sp. Goonoowigall in a well watered, forested area to the north.
The addition of a third New Caledonian species, S. procera, does not resolve relationships among them, suggesting rapid radiation after a dispersal event. Although the molecular analysis does not resolve the New Caledonian species, they are accepted as distinct taxa in the Flore de la Nouvelle-Calédonie et Dépendances (Dawson 1992, pp. 10–24), on the basis of leaf, flower and fruit characters. This analysis again showed that the likely progenitor of the New Caledonian lineage lies within the northern Queensland lineage sampled. The dispersal event is evidently later than the date of divergence of the two subclades and the branch lengths are suggestive of a date in the middle to late Miocene, or perhaps even early Pliocene.
The molecular analysis clearly showed that the plant from the Goonoowigall State Conservation Area is strongly supported as sister to S. cunninghamii, which both occur on the western slopes of New South Wales. This sister relationship was somewhat surprising because the leaf morphology of the new taxon did not fit what had seemed a plausible hypothesis, that the irregular leaf margins of S. cunninghamii and S. crenulata were a synapomorphy for this species pair. On the basis of the phylogeny and the morphological divergence, the Goonoowigall taxon is described below as S. whitei.
Taxonomy
Sannantha whitei Peter G.Wilson, sp. nov.
(Fig. 2)
Diagnosis
This species can be distinguished from other New South Wales species by its short, narrow, shortly acuminate leaves and few-flowered inflorescences.
Type: New South Wales, North Western Slopes: Goonoowigall State Conservation Area, near Goonoowigall Falls, Peter G. Wilson 1851 & T.C. Wilson, 6 Mar. 2013. (holotype: NSW; isotypes: BRI, CANB, NE).
Description
Spreading shrub 0.6–1 m high. Bark grey, persistent, fibrous. Branchlets somewhat quadrangular, ± flanged (more conspicuously so on older branchlets), margins entire; oil glands present, not markedly papillose; colleters (very small stipule-like structures) often occur in the axils of leaves. Leaves petiolate; petiole 0.5–0.6 mm long; lamina narrowly elliptical to narrowly obovate, 2.7–4.0 mm long, 0.7–1.1 mm wide, ± concolorous, concavo-convex, not keeled, oil glands prominent, particularly on lower surface, midrib not visible, intramarginal vein not visible, apex shortly acuminate, ~0.1 mm long. Inflorescence axillary, 1–3-flowered; peduncles 1.2–3.5 mm long; pedicels 1.2–3.0 mm long; bracts caducous. Flowers 5-merous (rarely 4-merous); hypanthium smooth, glandular, obconica1, 1.5–2.0 mm long, fused to the ovary throughout; sepals variable in size, up to ~0.5 × 1.2–1.3 mm; inner lobe (when developed) thin with irregular margins; outer lobe rudimentary or sometimes well developed, 0.2–0.5 mm long. Corolla 6.0–8.0 mm across; petals broadly suborbicular, 2.2–3.0 × 2.1–2.9 mm, white, oil glands present, margins somewhat irregular. Stamens (5–)6–8, in groups of 1 or 2 opposite the calyx lobes but towards their margins; filaments terete, 0.8–1.0 mm long, geniculate, with brown connective gland fused to upper part of filament at the bend; anthers adnate, dehiscing by pores, with loculi fused. Style terete, up to 1.0 mm long after anthesis, set into a pit on the ovary summit; stigma broadly capitate, ~0.5 mm wide. Ovary 3-locular; ovules 11–15 per loculus, arranged radially around placenta attached towards the base of the ovary. Fruit hemispherical, 1.0–1.8 × 1.8–2.4 mm, valves at rim level or slightly exserted. Seeds D-shaped, 0.5–0.7 mm long, brown, with flat sides and rounded backs, minutely reticulate.
Distribution
At present, S. whitei is known to occur only within the Goonoowigall State Conservation Area, south-east of Inverell. However, it is possible that the species might occur on parts of Middle Creek and its tributaries that are outside the present boundaries of the State Conservation Area.
Habitat
The species is locally common in damp places where it grows in sandy alluvial soils derived from granite, large boulders of which are common in the vicinity. Plants near Middle Creek are found adjacent to woodland of Eucalyptus prava and Callitris endlicheri, whereas plants that occur on damp ground away from the main watercourse are found growing with grasses and sedges.
Notes
With its short, narrow leaves, Sannantha whitei is superficially most similar morphologically to S. bidwillii, a Queensland species, but it differs from that species in having concolorous leaves, a very different habit and a distinctive ecology. Among the narrow-leaved species from New South Wales, S. whitei can be distinguished readily by having leaves (lamina plus petiole) that are <4.5 mm long.
The inflorescences are frequently only one- or two-flowered, a similarity with the phylogenetically close but morphologically rather different S. cunninghamii. The number of ovules observed in this species (11–15 per loculus) was often lower than the range indicated (‘14 or more’) when the genus was described (Wilson et al. 2007).
On specimens of S. whitei, colleters (very small stipule-like structures) can often be seen in the axils of leaves (Fig. 2B, C), and are discernible at the apices of the peduncles where they are borne in the axils of the caducous bracteoles (Fig. 2J). In other genera of Myrtaceae, these structures have occasionally been interpreted as stipules (e.g. Weberling 1956, 1966; Craven 1987; Snow et al. 2003), sometimes with reservations. Recent work by da Silva et al. (2012) has demonstrated that these ‘stipules’ are colleters, confirming the view of van Steenis (1969), and they identified three different types, namely, conic, petaloid and euryform. In S. whitei, most colleters appear to be euryform but some broader ones approach the petaloid type.
Etymology: the species is named for Gordon J. White (1936–) who was the first to collect it, in December 1992. Gordon worked in the Department of Botany at the University of New England (UNE) from 1962 until his retirement in 1996. In the last decade of his service, he took particular interest in the NCW Beadle Herbarium and is credited with the successful securing of grant funding to begin databasing the collection.
Other specimens examined
Acknowledgements
We thank Lesley Elkan for the excellent illustration and Angela White for biographical information. The first author thanks Trevor Wilson for his assistance in the field and for his photographs, Betsy Jackes (JCT) for supplying a sample of S. papillosa, and Adrien Wulff (University of New Caledonia and Institut Agronomique néo-Calédonien) for facilitating collection of S. procera. Financial support for early stages of this project was provided by ABRS. The first author thanks l’Institut Agronomique néo-Calédonien for assistance with travel to New Caledonia.
References
Bean AR (1997) Reinstatement of the genus Babingtonia Lindl. (Myrtaceae, Leptospermoideae). Austrobaileya 4, 627–645.Bean AR (1999) A revision of the Babingtonia virgata (J.R.Forst. & G.Forst.) F.Muell. complex (Myrtaceae) in Australia. Austrobaileya 5, 157–171.
Craven LA (1987) A taxonomic revision of Calytrix Labill. (Myrtaceae). Brunonia 10, 1–138.
| A taxonomic revision of Calytrix Labill. (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
da Silva CJ, Barbosa LC, Marques AE, Baracat-Pereira MC, Pinheiro AL, Meira RMSA (2012) Anatomical characterisation of the foliar colleters in Myrtoideae (Myrtaceae). Australian Journal of Botany 60, 707–717.
| Anatomical characterisation of the foliar colleters in Myrtoideae (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Dawson JW (1992) ‘Myrtaceae–Leptospermoideae. Flore de la Nouvelle-Calédonie et Dépendances. Vol. 18.’ (Muséum National d’Histoire Naturelle: Paris)
Golenberg EM, Clegg MT, Durbin ML, Doebley J, Ma DP (1993) Evolution of a noncoding region of the chloroplast genome. Molecular Phylogenetics and Evolution 2, 52–64.
| Evolution of a noncoding region of the chloroplast genome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2cXhsF2js7s%3D&md5=4f4e241279ff16d1bb792f5bb573fa27CAS | 8081547PubMed |
Lucas EJ, Harris SA, Mazine FF, Belsham SR, Nic Lughadha EM, Telford A, Gasson PE, Chase MW (2007) Suprageneric phylogenetics of Myrteae, the generically richest tribe in Myrtaceae (Myrtales). Taxon 56, 1105–1128.
| Suprageneric phylogenetics of Myrteae, the generically richest tribe in Myrtaceae (Myrtales).Crossref | GoogleScholarGoogle Scholar |
Maddison WP, Maddison DR (2000) ‘MacClade 4: Analysis of Phylogeny and Character Evolution.’ (Sinauer Associates: Sunderland, MA)
Martin HA (2006) Cenozoic climatic change and the development of the arid vegetation in Australia. Journal of Arid Environments 66, 533–563.
| Cenozoic climatic change and the development of the arid vegetation in Australia.Crossref | GoogleScholarGoogle Scholar |
Nylander JAA (2004) MrModeltest: program, ver.2.3. (Distributed by the author, Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden) Available at http://www.abc.se/~nylander/mrmodeltest2/mrmodeltest2.html [Verified 8 January 2014]
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542.
| MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space.Crossref | GoogleScholarGoogle Scholar | 22357727PubMed |
Simmons MP, Freudenstein JV (2011) Spurious 99% bootstrap and jackknife support for unsupported clades. Molecular Phylogenetics and Evolution 61, 177–191.
| Spurious 99% bootstrap and jackknife support for unsupported clades.Crossref | GoogleScholarGoogle Scholar | 21703355PubMed |
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49, 369–381.
| Gaps as characters in sequence-based phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD38zntlKjtg%3D%3D&md5=28e47d46829cef3c7826a15c80c9a1c4CAS | 12118412PubMed |
Snow N, Guymer GP, Sawvel G (2003) Systematics of Austromyrtus, Lenwebbia, and the Australian species of Gossia (Myrtaceae). Systematic Botany Monographs 65, 1–95.
| Systematics of Austromyrtus, Lenwebbia, and the Australian species of Gossia (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Swofford DL (2002) ‘PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods).’ (Sinauer Associates: Sunderland, MA)
Thornhill AH, Popple LW, Carter RJ, Ho SY, Crisp MD (2012) Are pollen fossils useful for calibrating relaxed molecular clock dating of phylogenies? A comparative study using Myrtaceae. Molecular Phylogenetics and Evolution 63, 15–27.
| Are pollen fossils useful for calibrating relaxed molecular clock dating of phylogenies? A comparative study using Myrtaceae.Crossref | GoogleScholarGoogle Scholar | 22197806PubMed |
van Steenis CGGJ (1969) Reduction of the genus Kania Schltr. to Metrosideros (Myrtaceae). Blumea 16, 357–359.
Weberling F (1956) Untersuchungen über rudimentäre Stipeln bei den Myrtales. Flora 143, 201–218.
Weberling F (1966) Additional notes on the Myrtaceous affinity of Kania eugenioides Schltr. Kew Bulletin 20, 517–520.
| Additional notes on the Myrtaceous affinity of Kania eugenioides Schltr.Crossref | GoogleScholarGoogle Scholar |
Wilson PG (1991) Baeckea. In ‘Flora of New South Wales. Vol. 2’. (Ed. GJ Harden) pp. 181–186. (New South Wales University Press: Sydney, NSW)
Wilson PG, Heslewood MM, Quinn CJ (2007) Re-evaluation of the genus Babingtonia (Myrtaceae) in Eastern Australia and New Caledonia. Australian Systematic Botany 20, 302–318.
| Re-evaluation of the genus Babingtonia (Myrtaceae) in Eastern Australia and New Caledonia.Crossref | GoogleScholarGoogle Scholar |