Using population genetic tools to develop a control strategy for feral cats (Felis catus) in Hawai‘i
Heidi Hansen A B , Steven C. Hess C E , David Cole A D and Paul C. Banko CA Hawai‘i Cooperative Studies Unit (PACRC, UH Hilo), US Geological Survey Pacific Island Ecosystems Research Center, Kīlauea Field Station, PO Box 44, Hawai‘i National Park, HI 96718 USA.
B Current address: Department of Land and Natural Resources, Division of Forestry and Wildlife, 19 East Kawili Street, Hilo, HI 96720, USA.
C US Geological Survey Pacific Island Ecosystems Research Center, Kīlauea Field Station, PO Box 44, Hawai‘i National Park, HI 96718 USA.
D Current address: University of Hawai‘i at Mānoa, 3190 Maile Way, Room 101, Honolulu, HI 96822, USA.
E Corresponding author. Email: steve_hess@usgs.gov
Wildlife Research 34(8) 587-596 https://doi.org/10.1071/WR07043
Submitted: 10 April 2007 Accepted: 5 October 2007 Published: 18 December 2007
Abstract
Population genetics can provide information about the demographics and dynamics of invasive species that is beneficial for developing effective control strategies. We studied the population genetics of feral cats on Hawai‘i Island by microsatellite analysis to evaluate genetic diversity and population structure, assess gene flow and connectivity among three populations, identify potential source populations, characterise population dynamics, and evaluate sex-biased dispersal. High genetic diversity, low structure, and high number of migrants per generation supported high gene flow that was not limited spatially. Migration rates revealed that most migration occurred out of West Mauna Kea. Effective population size estimates indicated increasing cat populations despite control efforts. Despite high gene flow, relatedness estimates declined significantly with increased geographic distance and Bayesian assignment tests revealed the presence of three population clusters. Genetic structure and relatedness estimates indicated male-biased dispersal, primarily from Mauna Kea, suggesting that this population should be targeted for control. However, recolonisation seems likely, given the great dispersal ability that may not be inhibited by barriers such as lava flows. Genetic monitoring will be necessary to assess the effectiveness of future control efforts. Management of other invasive species may benefit by employing these population genetic tools.
Introduction
Population genetic tools have proven beneficial for the management and conservation of rare and endangered wildlife (Paetkau et al. 1997; Spong et al. 2000). Only recently have these tools been applied in invasive species management (Robertson and Gemmell 2004; Abdelkrim et al. 2005; Rollins et al. 2006). Invasive species are the main cause of species extinctions in island ecosystems (Courchamp et al. 1999) and the second main cause of biodiversity loss after habitat destruction (Vitousek et al. 1997). Successful management of invasive species requires identifying a target population of manageable size that has a low recolonisation risk (Robertson and Gemmell 2004). Attempts to control only a fraction of the population or a sink population could result in rapid recolonisation (Robertson and Gemmell 2004; Abdelkrim et al. 2005). Identifying routes of potential migration is difficult using direct observations, but vital for controlling invasive species (Robertson and Gemmell 2004; Abdelkrim et al. 2005; Rollins et al. 2006). Population genetics can provide valuable information about the demographic status and dynamics of invasive species and may provide an alternative approach for developing control strategies (Robertson and Gemmell 2004; Abdelkrim et al. 2005; Pontier et al. 2005).
Feral cats (Felis catus) are currently listed as one of the ‘100 world’s worst invasive alien species’ (Lowe et al. 2000). Domestic cats were brought to Hawai‘i on European ships in the late 1700s (King 1984) and feral animals were reported by 1840 in remote montane areas of Hawai‘i Island (Brackenridge 1841). Currently, feral cats occur in low densities in montane forests and subalpine areas of Maui (Simons 1983) and Hawai‘i Island (Hu et al. 2001) and are frequent predators of endangered Hawai‘ian birds including colonial seabirds (Smith et al. 2002), ground-nesting waterfowl (Banko 1992), and tree-nesting passerines (Hess et al. 2004). Cats also carry Toxoplasma gondii, which has caused fatal toxoplasmosis in endangered Hawai‘ian birds (Work et al. 2000, 2002) and Hawai‘ian monk seals (Monachus schauinslandi) (Honnold et al. 2005).
The behaviour of feral cats in remote subalpine and alpine environments of Hawai‘i makes traditional methods to understand population dynamics problematic; they are solitary, elusive, hard to capture, and inhabit areas that are difficult to survey (Hess et al. 2004). Their dispersal patterns and the inaccessibility of remote locations also make them logistically difficult to manage. Research on the genetic structure of feral cats in island ecosystems is rare (Pontier et al. 2005), but can provide valuable information for formulating control strategies and determining the scale and location of control efforts (Rollins et al. 2006). Here, we describe the use of seven highly polymorphic microsatellite markers to estimate the genetic structure of three feral cat populations on Hawai‘i Island. The objectives of our research were to (1) evaluate genetic diversity and population structure, (2) assess levels of gene flow and connectivity between populations, (3) identify potential source populations, (4) characterise population dynamics and (5) evaluate evidence for sex-biased dispersal. We present results that can be used to formulate an island-wide control strategy.
Study areas
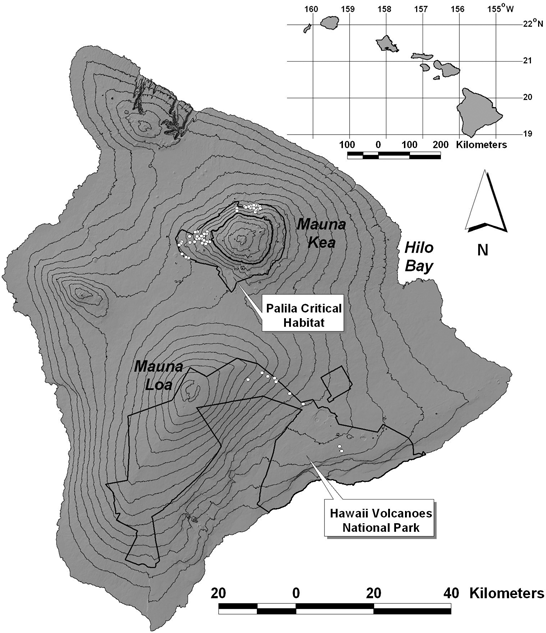
Study areas were located on Hawai‘i Island in Hawai‘i Volcanoes National Park (HAVO), and on North Mauna Kea (NMK), and West Mauna Kea (WMK) in designated critical habitat for palila (Loxioides bailleui) (Fig. 1). NMK and WMK were characterised as dry subalpine woodlands from 1701 to 2835 m elevation. HAVO extended from 800 to 2700 m elevation and consisted of montane wet forest grading into drier ‘ōhi‘a (Metrosideros polymorpha) scrub and subalpine shrubland. Substrates in HAVO were geologically young and interspersed with recent lava flows. HAVO and Mauna Kea were separated by extensive recent lava flows. The linear distances separating WMK from NMK, WMK from HAVO, and NMK from HAVO were 18 km, 50.2 km, and 53.5 km, respectively, and greater than the average home ranges reported for feral cats on Mauna Kea (1418 ha and 770 ha, respectively: D. Goltz, unpubl. data). The approximate areas from which cats were captured varied from 8 km2 at NMK and 32 km2 at WMK, to 87 km2 at HAVO. Traps were arranged on transects perpendicular to elevation contours on NMK and WMK, and parallel to contours in HAVO. The population density of cats was unknown.
|
Methods
Population sampling and microsatellite analysis
Feral cats were trapped during 2000–05 to reduce predation on endangered species such as nēnē (Hawai‘ian goose, Branta sandvicensis) and ‘Ua‘u (Hawai‘ian petrel, Pterodroma sandwichensis) in HAVO, and palila on NMK and WMK. Traps were checked daily and cats were euthanised according to University of Hawai‘i IACUC Protocol 97-063. We collected muscle tissue samples from 85 feral cats (49 males, 36 females) and stored samples in lysis buffer (0.1 m Tris–HCl pH 8.0, 0.1 m sodium EDTA, 2% SDS) at –20°C until extraction. We extracted genomic DNA from the tissue using the QIAGEN® DNeasyTM Tissue Kit (Qiagen, Inc., Valencia, CA, USA).
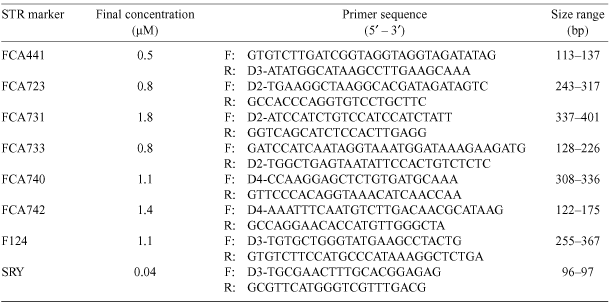
We generated complete genotypes for all feral cat samples using seven polymorphic short-tandem repeat (STR) microsatellite markers previously developed and optimised for multiplexing of domestic cats (Menotti-Raymond et al. 2005) and included a gender-identifying sequence tagged site (STS) from the domestic cat Y-chromosome SRY gene. The markers were labelled with D2, D3 and D4 WellRED fluorescent dyes (Beckman Coulter, Fullerton, CA, USA) at the five-prime end of either the forward or reverse primers. This allowed multiplexing of all eight marker loci in one sequence run by distinguishing loci and associated alleles that overlapped in size.
A single PCR (polymerase chain reaction) was performed for each sample in a 20.0-µL volume containing 1 × PCR Gold buffer (Applied Biosystems, Fullerton, CA, USA), with final reaction concentrations (adapted from Menotti-Raymond et al. 2005) as follows: 1.5 mm MgCl2, 0.8 mm dNTPs, 1 unit Amplitaq gold DNA polymerase (Applied Biosystems), 0.16 mg mL–1 bovine serum albumin, 8 µL microsatellite primer mix (see final concentrations in Table 1), and ~20 ng genomic DNA. PCRs were performed with a MJ Research PTC-200 DNA thermocycler, using conditions optimised by Menotti-Raymond et al. (2005). The eight-primer pair multiplexes were then visualised on a Beckman Coulter CEqn 8000 automated capillary sequencer, one lane for each DNA sample (Core Genetics Facility, University of Hawai‘i at Hilo). Allele sizes were estimated using CEqn 8000 version 7.0 and then visually inspected, taking into consideration the expected allele size in base pairs for each of the eight loci and the original DNA clones from which the microsatellite loci were developed (Menotti-Raymond et al. 2005).
|
Genetic diversity
Genetic diversity for each population was summarised as the average number of alleles per locus (A) and average observed (HO) and expected (HE) heterozygosities (Nei 1978) with Fstat 2.9.3 (Goudet 2001). We calculated the unbiased inbreeding coefficient f (similar to Fis: Weir and Cockerham 1984) for each population–locus combination and tested for deviations from Hardy–Weinberg equilibrium with Genepop 3.4 (Raymond and Rousset 1995). Departure from Hardy–Weinberg expectations was assessed by exact tests with unbiased P-values based on a Markov chain simulation (with 1000 as dememorisation number, 500 batches and 1000 iterations per batch: Guo and Thompson 1992). Loci were combined by Fisher’s method (Raymond and Rousset 1995) to examine departure from equilibrium for each population. A Bonferonni correction was used to adjust significance levels across multiple tests (Goudet 2001). All loci occurred on different chromosomes or different linkage groups on the same chromosome (Menotti-Raymond et al. 2005) and were considered independent markers.
Population structure
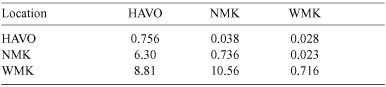
We investigated population structure among the three populations by using several methods with analyses for all feral cats, and separate analyses for males and females. We calculated the unbiased estimator θ (analogous to FST: Weir and Cockerham 1984) for all population pairs with Fstat (Goudet 2001). In addition to θ, ρ (analogous to RST: Slatkin 1995) was estimated with RST Calc 2.2 (Goodman 1997) after standardisation of allele sizes in RST Standardise 2.2 (Goodman 1997). We tested for correlation between the values of θ and ρ by using a Mantel test (Pearsons correlation, with 10000 iterations: Manly 1991) conducted in Mantel 2.0 (Liedloff 1999). We tested the significance levels of θ and ρ for each population pair by calculating P-values and 95% confidence intervals (CI) by a bootstrap procedure. We compared pairwise θ estimates with log-transformed geographic distances (ln(km)) to assess whether dispersal is limited spatially for feral cats. Significance of the relationships was tested by a Mantel test (Pearsons correlation, with 10000 iterations: Manly 1991) conducted in Mantel 2.0 (Liedloff 1999). Geographical distances between individuals were calculated with ArcView GIS 3.2 (ESRI 1999).
We used a Bayesian Markov chain Monte Carlo (MCMC) approach to cluster individual genotypes from all cats (n = 85), male cats (n = 49) and female cats (n = 36) into respective populations and determine the most likely number of populations (K) as implemented in the program Structure 2.0 (Pritchard et al. 2000). Posterior probability values for K (log-likelihood; ln(L)) were estimated for preassigned number of clusters (K = 1–6 for all cats, K = 1–3 for males and females) using the mixed ancestry (admixture) model, a burn-in of 30 000 iterations (checking that parameters α and likelihood had converged), and 100 000 MCMC repetitions for 3–4 independent runs. We used only genetic information and excluded geographic location from analyses. The K value where the likelihood reached an asymptote was chosen as the number of populations (Pritchard et al. 2000).
We then conducted individual-based assignment tests that assign an individual to the population in which its genotype is most likely to occur to identify possible migrants (Aspi et al. 2006). First, we assigned samples into respective populations based on the highest proportion of membership (q) from results obtained in Structure. Second, we used the Rannala and Mountain (1997) Bayesian individual assignment method to estimate the likelihood that a cat originated from a given population as implemented in Geneclass2 (Piry et al. 2004). The probability of an individual being a resident was compared with randomly generated genotypes (10 000 replicates) and an individual was rejected from the population if the value was below P < 0.01
Effective population size
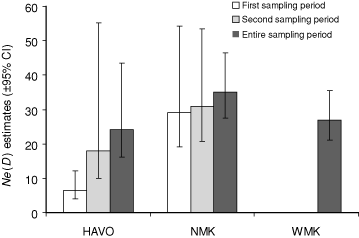
To estimate the current number of successfully breeding individuals per population, we used the method of linkage disequilibrium described by Bartley et al. (1992) to estimate the effective population size (Ne(D)) and 95% CI for each population as implemented in NeEstiMator (Peel et al. 2004). To investigate short-term trends of Ne(D), we identified temporal sampling periods that corresponded to trapping intervals in HAVO and NMK and estimated the Ne(D) and 95% CI for each sampling period by Hill’s (1981) one-sample method. Owing to the small numbers of cats caught each year in HAVO, the first and second temporal sampling periods occurred from 2000 to 2002 (n = 6) and 2003 to 2005 (n = 9), respectively, with samples pooled to increase sample sizes. The first and second sampling periods for NMK occurred in 2004 (n = 18) and 2005 (n = 21), respectively. This analysis was not available for WMK as there was only one sampling period, in 2003 (n = 31).
Gene flow and migration rate
To obtain indirect measures of gene flow between populations, we estimated the number of migrants per generation (Nm), where N is the effective population size, m is the proportion of migrants per generation, and
Nm = (1/FST – 1)/4
(Slatkin 1995). Nm estimates are based on historical rates of gene flow and include only individuals that successfully reproduce (Pearse and Crandall 2004).
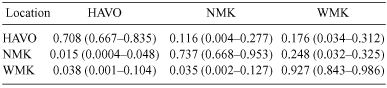
To determine possible source populations that could be targeted for control (Rollins et al. 2006), we estimated recent migration rates among populations by the Bayesian approach as implemented in Bayesass+ 1.3 (Wilson and Rannala 2003) and approximated 95% CI. The program was run with a MCMC length of 3 000 000 iterations, a burn-in period of 100 000 (checking that the chains had converged and the log-likelihood values had peaked), and the input parameters (ΔP = allele frequency, Δm = migration, Δ f) set at 0.15, 0.10, and 0.20, respectively. This analysis included all migrants regardless of success at reproduction (Rollins et al. 2006).
Sex-biased dispersal
To assess sex-biased dispersal, we examined potential differences between sexes in genetic structure and relatedness. We performed an assignment t-test in Fstat as described by Goudet et al. (2002) and calculated mean assignment indices (mAIc), θ estimates and f estimates for females and males among the populations (Favre et al. 1997). We used program Identix 4.03 (Belkhir et al. 2002) to estimate pairwise relatedness (rxy) of males and females by applying the method of Lynch and Ritland (1999) and approximated 95% CI. We compared the relatedness between female and male cats within individual populations and relatedness of females among all populations. To investigate potential differences in relatedness between populations, we examined rxy estimates and geographic distances between individuals by the Mantel test (Pearsons correlation, with 10 000 iterations: Manly 1991) conducted in Mantel 2.0 (Liedloff 1999).
Results
Genetic diversity
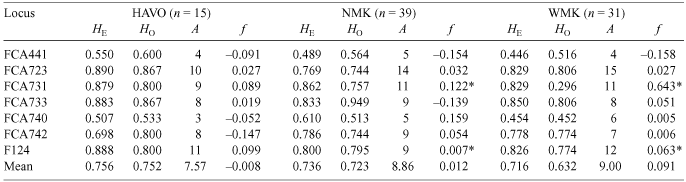
The mean number of alleles (A) ranged from 7.57 ± 2.99 (mean ± s.d.) to 9.00 ± 3.83 according to population (Table 2). Mean HO ranged from 0.30 to 0.95 according to locus, and ranged from 0.63 ± 0.03 (WMK) to 0.76 ± 0.04 (HAVO) according to population. Mean HE and HO were not significantly different in any population but mean HE was larger than HO (0.72 ± 0.07) in WMK. Except for F124 and FCA731 (P < 0.002) in NMK and WMK, the single-locus f values did not differ from zero (P > 0.05). Multilocus f values ranged from –0.01 (P = 0.16) in HAVO to 0.09 (P < 0.0007) in WMK with an overall value of 0.03 (P = 0.0001) between all loci and populations. After Bonferonni corrections, f values showed a significant departure from Hardy–Weinberg equilibrium in NMK and WMK, suggesting that either (1) inbreeding may have increased, we unintentionally sampled parent–offspring pairs, or (2) null alleles are present in these populations (Kyle and Strobeck 2001; Schwartz et al. 2003; Aspi et al. 2006).
Population structure
Overall estimates of ρ (0.023; P = 0.008) showed results similar to those of θ (0.028; P = 0.0001). Pairwise θ estimates were all significant (P < 0.05) and calculated (with 95% CI) as 0.038 (HAVO–NMK, 0.008–0.071), 0.028 (HAVO–WMK, 0.013–0.041) and 0.023 (NMK–WMK, 0.006–0.043). Pairwise ρ-values (with 95% CI) were significant (P < 0.05) for HAVO–WMK and NMK–WMK (0.047, 0.017–0.135; and 0.025, 0.008–0.083, respectively) but not for HAVO–NMK (0.004, P = 0.274, –0.009–0.084). Pairwise values of ρ and θ were not correlated (Mantel test r = –0.649, P = 0.853) and pairwise ρ-values had extremely large 95% CI (almost twice as large as θ estimates). When populations are weakly structured with high rates of gene flow, θ provides a more accurate estimator than ρ (Balloux and Goudet 2002). Therefore, ρ-values were not used in further analyses. Genetic differentiation between population pairs was not significantly correlated with distance (ln(km)) (Mantel test r = 0.789, P = 0.167).
Bayesian clustering of all feral cats suggested the presence of three clusters (ln(L) = –2037; not shown) with membership in each cluster ranging from q = 0.35 to q = 0.95 (mean q = 0.75). Females were split into two clusters (ln(L) = –818) with membership ranging from q = 0.59 to q = 0.98 (mean q = 0.88). Female cats from HAVO and NMK created one cluster while WMK females created another. For males, a single cluster (K = 1) was the most likely (ln(L) = –1261).
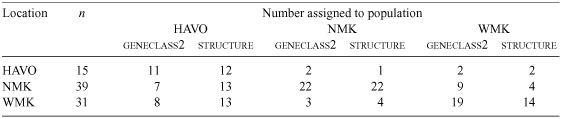
Assignment tests correctly assigned most individuals to the area in which they were trapped (Geneclass2: 62.4%; Structure: 56.4%). Considering the two closest populations (18 km apart), 23.1% (Geneclass2) and 10.3% (Structure) of individuals captured in NMK were assigned to WMK. Of those misassigned in WMK, 25.8% (Geneclass2) and 41.9% (Structure) were assigned to HAVO (Table 3). In Geneclass2, two individuals were identified as samples that could not be grouped into a population, and thus were likely to be migrants from an unsampled population (Cegelski et al. 2003). Another two individuals were identified in both tests as being likely migrants (probability of assignment (P) >0.80) from WMK into both HAVO and NMK.
|
Effective population size
The overall Ne(D) estimates for HAVO, NMK, and WMK (with 95% CI) were 24.2 (19.2–54.2), 35 (27.5–46.4), and 26.9 (21.2–35.4), respectively. In HAVO, the Ne(D) estimates from 2000–02 samples (n = 6) and 2003–05 samples (n = 9) increased from 6.4 (4.0–12.2) to 18 (10.0–55.3), respectively. In NMK, the Ne(D) estimates from 2004 samples (n = 18) and 2005 samples (n = 21) increased from 29.2 (19.1–54.2) to 30.8 (20.8–53.5), respectively. Although not significant, these estimates may suggest population growth for HAVO and NMK (Fig. 2).
|
Gene flow and migration rate
Nm estimates suggested high gene flow between feral cat populations with the effective number of migrants per generation ranging between 6.3 and 10.6 (Table 4). Number of migrants per generation was lowest between HAVO and NMK and highest in the adjacent populations on Mauna Kea. The mean posterior probabilities of migration rates showed that most individuals were native to their capture locations in all populations, with the most originating in WMK (0.708–0.927) (Table 5). There was a relatively high degree of migration between populations from WMK to NMK (m = 0.248; 95% CI = 0.032 – 0.325) and HAVO (m = 0.176; 95% CI = 0.034–0.312). In contrast, migration rates from both HAVO and NMK into the other populations were very low, with the smallest migration rate from HAVO to WMK (m = 0.015; 95% CI = 0.0004–0.048).
|
|
Sex-biased dispersal
Assignment t-test results supported male-biased dispersal and female philopatry. Relatedness of males and females differed significantly (P < 0.05) and the mAIc of males among populations was significantly lower than that of females (mAIc = –0.900, 1.23, P = 0.007, respectively). The estimate of f was significantly higher for males (F = 0.085) than for females (F = –0.007) (P = 0.008) and the average θ estimate across the populations for males (θ = 0.013) was significantly lower than that calculated for females (θ = 0.053) (P = 0.028).
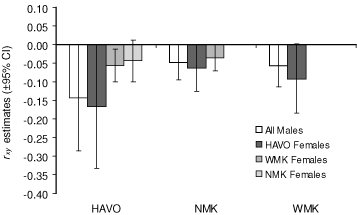
We found no differences in relatedness between female and male cats within and among populations (Fig. 3). Mean rxy values suggested little to no relatedness within and among populations (as shown by the negative estimates) and both male and female cats in HAVO were less related to each other than to cats in other populations, most likely owing to the larger sampling area of HAVO compared with the other populations. Among populations, pairwise rxy values were significantly (P < 0.0001) correlated with geographic distance, having a greater inverse correlation for female cats (Mantel test r = –0.161) than for all cats (Mantel test r = –0.131) and very little inverse correlation for male cats (Mantel test r = –0.095). Within populations, rxy values and distance were not correlated for male cats (P > 0.5 in all populations). For females, rxy values declined significantly with distance in HAVO and WMK (HAVO, Mantel test r = –0.576, P = 0.005; WMK, Mantel test r = –0.386, P = 0.002), although not in NMK (Mantel test r = –0.05, P = 0.587).
|
The most related individuals (rxy = 0.86), an adult and a juvenile female, were captured within 15 days of each other at a distance of 1.4 km in WMK. It is likely that this was a mother–daughter pair. The most closely related individuals (rxy = 0.62) separated by the greatest distance (65.9 km) were two females captured in HAVO and NMK and were likely siblings or a mother–daughter pair.
Discussion
Genetic structure suggests that feral cats on Hawai‘i Island exhibit long-distance dispersal between populations and their movements are not inhibited by current control efforts or barriers such as extensive lava flows. As expected, given their European ancestry, the genetic diversity of feral cats we examined in Hawai‘i (A = 7.57–9.00, HO = 0.70) was similar to that of European domestic cats (A = 14.2, HO = 0.70: Pierpaoli et al. 2003) but was greater than that reported for feral cats recently introduced from France to a subantarctic island (A = 3.67–7.00, HO = 0.53: Pontier et al. 2005) and also for cat colonies in France (A = 4.38–7.78, HO = 0.61) that experience low dispersal rates owing to barriers such as heavy-traffic roads (Say et al. 2003). We also found very little genetic differentiation (θ = 0.028) between populations and dispersal was not spatially limited. Generally, FST values below 0.05 suggest high levels of gene flow and population connectivity (Thulin et al. 2006). Therefore, we conclude that the three populations were not founded independently, there was rapid colonisation from initial founders owing to high growth rates, and ongoing gene flow has occurred (Abdelkrim et al. 2005; Pontier et al. 2005). Apparently, feral cats in Hawai‘i have not differentiated markedly from their initial founders (Pontier et al. 2005). Although we have no definitive evidence to rule out the possibility that domestic house cats were recruited into nearby feral populations, all of the cats we captured lacked diversity in coat coloration and had reverted back to pelage characteristics similar to those of European wildcats (Felis silvestris) (Lowe et al. 2000; Beaumont et al. 2001).
We found some evidence of inbreeding in the Mauna Kea populations (f > 0.09) and between all populations (F = 0.03), although this was low compared with estimates of isolated feral cat populations in urban France (F = 0.14) (Say et al. 2003) and on a subantarctic island (f > 0.11) (Pontier et al. 2005). Inbreeding may be the result of factors that cause low population density, such as kitten mortality due to feline leukemia virus, feline immunodeficiency virus, and toxoplasmosis (Molsher 1999), all of which have been documented in feral cats on Mauna Kea (Danner et al. 2007). Inbreeding may also reduce fitness and resistance to diseases (Coltman et al. 1999), mating success (Slate et al. 2000) and juvenile survival (Coltman et al. 1998).
Despite the positive inbreeding coefficients, WMK and NMK, separated by the shortest distance (18 km), had high levels of gene flow between them, with 10.6 cats migrating per generation. Kaeuffer et al. (2004) estimated that the mean generation time for feral cats in France was 3.38 years. This suggests that >10 cats migrated between WMK and NMK and successfully reproduced in 3 years. Given the home-range sizes of feral cats reported from similar environments in Australia (Edwards et al. 2001), it is not surprising that cats disperse long distances. Often populations in close proximity receive a greater number of migrants than populations farther apart (Whitlock and McCauley 1999), and, accordingly, less gene flow occurred between Mauna Kea and HAVO. It is surprising, though, that an estimated 6.3–8.8 cats per generation may have migrated between these populations given the harsh environment between these areas. However, our estimates of indirect gene flow are presented only as an index of relative measures of connectivity between populations and some assumptions of this analysis may have been violated (Whitlock and McCauley 1999; Cegelski et al. 2003).
As for other carnivores, the spatial organisation of cats is determined by the abundance and distribution of available prey and receptive mates and the proximity to human habitation (Pontier et al. 1995). The density of cats may be higher on Mauna Kea, particularly WMK, than in HAVO owing to abundant prey (Hess et al. 2007), which may facilitate increased reproduction, survival, and dispersal to other locations. Accordingly, migration rates showed that most dispersing cats originated and immigrated from WMK into the other populations with little migration occurring into WMK. Also, only 45–61% of individuals captured in WMK were correctly assigned to their origin. The ‘misassigned’ individuals were likely migrants from an unsampled population or offspring of migrants (Cegelski et al. 2003; Rollins et al. 2006). In fact, two migrants were captured in both HAVO and NMK and assigned to WMK. This suggests that cats may not be able to easily integrate with some residents and may disperse long distances to fill empty territories created by current predator-removal efforts. Also, cats may disperse more during the seabird breeding season to prey upon these burrow-nesting endangered species (Hu et al. 2001).
Mauna Kea populations had significantly more successfully breeding individuals than did HAVO, as shown in the larger Ne estimates (DeYoung and Honeycutt 2005). Because we trapped consistently between 2000 and 2005, our samples had overlapping generations and immediate evaluation of Ne would have been difficult using more common temporal methods that require at least two non-overlapping generations (Waples 1989). Bartley’s linkage disequilibrium method (Bartley et al. 1992) has an advantage in that population trends can be evaluated (Leberg 2005) and only one sampling period is required regardless of generation length (Bartley et al. 1992). Both HAVO and NMK apparently experienced population expansion with increasing Ne estimates, indicating insufficient control efforts between trapping periods. However, our sample sizes were small and our results should be used only to evaluate control strategies, as sample sizes over 90 may be necessary to obtain precise estimates of Ne when using the linkage disequilibrium method (Bartley et al. 1992). Also, Mauna Kea populations may be experiencing introgression from adjacent unsampled populations which would explain both the increase in Ne estimates and misassigned individuals (Roman and Palumbi 2003; Spencer and Hampton 2005).
Although our results support high gene flow, population structuring was evident in the Bayesian clustering results that showed three clusters of cats. Cats captured in HAVO had a greater proportion of individuals correctly assigned to that population whereas individuals in Mauna Kea had more ambiguous assignments. The discrepancies between genetic structure and assignment tests could be a result of several reasons such as the distance and terrain separating populations, predator-control efforts reducing encounter rates, and sampling individuals that have mixed or indistinct ancestry (Cegelski et al. 2003). When analysing individual sexes, population structuring suggested two clusters of females and only one of males. Also, assignment t-tests and relatedness estimates supported male-biased dispersal, which is common in many mammal species (Goudet et al. 2002). More unusual was the similar relatedness we found between males and females. However, relatedness of females, but not males, decreased significantly with geographic distance, further supporting female philopatry. Similar to other research, we found that sex-biased dispersal estimates derived using genetic-based assignment techniques mirrored radio-telemetry findings, and did not require as much field effort (DeYoung and Honeycutt 2005). Goudet et al. (2002) suggested that short-distance dispersal may be sex-biased to avoid inbreeding. Alternatively, long-distance dispersal may be a means to colonise empty patches and is unlikely to be sex-biased. In fact, Spong and Creel (2001) reported that ~20% of female lions (Panthera leo) in Africa emigrate to new territories. This is consistent with our finding that the most closely related individuals separated by the furthest distance were two females.
Our results can be used to design an effective plan for feral cat control. Although complete eradication of feral cats has occurred on several small islands (Nogales et al. 2004), Hawai‘i Island would require large-scale control programs, which are typically difficult to implement (Pontier et al. 2005). One major limitation for control efforts is the ability of the invasive species to disperse from neighbouring populations and recolonise (Abdelkrim et al. 2005). The genetic structure of feral cats in Hawai‘i indicates great dispersal ability, therefore control will be very difficult and recolonisation seems highly likely. The reproductive biology and life history of the invasive species can also determine the ease of control and potential for recolonisation (Myers et al. 2000). Female cats reach sexual maturity between 6 and 8 months and males between 8 and 10 months and can breed 2–3 times a year. However, reproduction can be delayed (Jones and Coman 1982; Say et al. 1999) as shown by Bester et al. (2002), who reported that a drastic decrease of feral cats following eradication efforts on subantarctic Marion Island caused a decline in pregnancy rates and fecundity, possibly owing to a lower encounter rate between the sexes. Control efforts need to minimise gene flow to no more than one migrant per generation in order to decrease genetic diversity (Wright 1969) and thus create small or fragmented populations in which inbreeding is more likely to occur. Targeting the source population for control is important when the animal disperses long distances on unknown pathways (Rollins et al. 2006). On Hawai‘i Island, control efforts on Mauna Kea may benefit endangered wildlife at other locations by reducing dispersing feral cats. Also, the sex that disperses the most could be targeted for control (Rollins et al. 2006); however, we do not recommend targeting male feral cats as both sexes exhibited some dispersal.
To minimise risk of failure in controlling invasive species and to reduce management cost and loss of native species, genetic monitoring of the invasive species should be a preliminary step in the management process in order to assess the effectiveness of control efforts (Robertson and Gemmell 2004; Abdelkrim et al. 2005; Rollins et al. 2006). For example, a decrease in genetic diversity, increase in inbreeding, and decrease in effective population size would all indicate successful control strategies (DeYoung and Honeycutt 2005). Assignment tests can be used to identify captured individuals as either control survivors or recolonisers following control efforts (Robertson and Gemmell 2004). Management of other invasive vertebrates may benefit by employing these population genetic tools.
Acknowledgements
This project was funded in part by the USGS–NPS Natural Resources Partnership Program (NRPP) and the USGS Invasive Species Program. We thank D. Hu, K. Misajon, R. Swift, J. T. Tunison, D. M. Goltz, R. M. Danner, D. Nelson, and R. M. Stephens for assistance, facilitation, guidance, and samples. N. Seavy and two anonymous reviewers provided helpful comments. Finally, we thank our research interns E. Baldwin and A. Bies, who provided invaluable assistance in gathering data. Any use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the USA Government.
Abdelkrim, J. , Pascal, M. , Calmet, C. , and Samadi, S. (2005). Importance of assessing population genetic structure before eradication of invasive species: examples from insular Norway rat populations. Conservation Biology 19, 1509–1518.
| Crossref | GoogleScholarGoogle Scholar |
Bester, M. N. , Bloomer, J. P. , Van Aarde, R. J. , Erasmus, B. H. , Van Rensburg, P. J. J. , Skinner, J. D. , Howell, P. G. , and Naude, T. W. (2002). A review of the successful eradication of feral cats from the sub-Antarctic Marion Island, southern Indian Ocean. South African Journal of Wildlife Research 32, 65–73.
Cegelski, C. C. , Waits, L. P. , and Anderson, N. J. (2003). Assessing population structure and gene flow in Montana wolverines (Gulo gulo) using assignment-based approaches. Molecular Ecology 12, 2907–2918.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
Favre, L. , Balloux, F. , Goudet, J. , and Perrin, N. (1997). Female-biased dispersal in the monogamous mammal Crocidura russula: evidence from field data and microsatellite patterns. Proceedings of the Royal Society of London. Series B. Biological Sciences 264, 127–132.
| Crossref | GoogleScholarGoogle Scholar |
Goudet, J. , Perrin, N. , and Waser, P. (2002). Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Molecular Ecology 11, 1103–1114.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
Kyle, C. J. , and Strobeck, C. (2001). Genetic structure of North American wolverine (Gulo gulo) populations. Molecular Ecology 10, 337–347.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
Lynch, M. , and Ritland, K. (1999). Estimation of pairwise relatedness with molecular markers. Genetics 152, 1753–1766.
| PubMed |
Menotti-Raymond, M. A. , David, V. A. , Wachter, L. L. , Butler, J. M. , and O’Brien, S. J. (2005). An STR forensic typing system for genetic individualization of domestic cat (Felis catus) samples. Journal of Forensic Sciences 50, 1061–1070.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
Myers, J. H. , Simberloff, D. , Kuris, A. M. , and Carey, J. R. (2000). Eradication revisited: dealing with exotic species. Trends in Ecology & Evolution 15, 316–320.
| Crossref | GoogleScholarGoogle Scholar |
Pierpaoli, M. , Birò, Z. S. , Herrmann, M. , Hupe, K. , Fernández, M. , Ragni, B. , Szemethy, L. , and Randi, E. (2003). Genetic distinction of wildcat (Felis silvestris) populations in Europe, and hybridization with domestic cats in Hungary. Molecular Ecology 12, 2585–2598.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
Slate, J. , Kruuk, L. E. B. , Marshall, T. C. , Pemberton, J. M. , and Clutton-Brock, T. H. (2000). Inbreeding depression influences lifetime breeding success in a wild population of red deer (Cervus elaphus). Proceedings of the Royal Society of London. Series B. Biological Sciences 267, 1657–1662.
| Crossref | GoogleScholarGoogle Scholar |