Finding genes for economically important traits: Brahman cattle puberty
M. R. S. Fortes B C , S. A. Lehnert B , S. Bolormaa D , C. Reich D , G. Fordyce E , N. J. Corbet B , V. Whan B , R. J. Hawken B and A. Reverter B FA Cooperative Research Centre for Beef Genetic Technologies.
B CSIRO Livestock Industries, Queensland Bioscience Precinct, St Lucia, Qld 4067, Australia.
C The University of Queensland, School of Veterinary Science, Gatton, Qld 4343, Australia.
D Biosciences Research Division, Department of Primary Industries Victoria, 1 Park Drive, Bundoora, Vic. 3083, Australia.
E Queensland Alliance for Agriculture and Food Innovation, Centre for Animal Science, The University of Queensland, St Lucia, Qld 4067, Australia.
F Corresponding author. Email: Tony.Reverter-Gomez@csiro.au
Animal Production Science 52(3) 143-150 https://doi.org/10.1071/AN11165
Submitted: 5 August 2011 Accepted: 10 February 2012 Published: 6 March 2012
Journal Compilation © CSIRO Publishing 2012 Open Access CC BY-NC-ND
Abstract
Age at puberty is an important component of reproductive performance in beef cattle production systems. Brahman cattle are typically late-pubertal relative to Bos taurus cattle and so it is of economic relevance to select for early age at puberty. To assist selection and elucidate the genes underlying puberty, we performed a genome-wide association study (GWAS) using the BovineSNP50 chip (~54 000 polymorphisms) in Brahman bulls (n = 1105) and heifers (n = 843) and where the heifers were previously analysed in a different study. In a new attempt to generate unbiased estimates of single-nucleotide polymorphism (SNP) effects and proportion of variance explained by each SNP, the available data were halved on the basis of year and month of birth into a calibration and validation set. The traits that defined age at puberty were, in heifers, the age at which the first corpus luteum was detected (AGECL, h2 = 0.56 ± 0.11) and in bulls, the age at a scrotal circumference of 26 cm (AGE26, h2 = 0.78 ± 0.10). At puberty, heifers were on average older (751 ± 142 days) than bulls (555 ± 101 days), but AGECL and AGE26 were genetically correlated (r = 0.20 ± 0.10). There were 134 SNPs associated with AGECL and 146 SNPs associated with AGE26 (P < 0.0001). From these SNPs, 32 (~22%) were associated (P < 0.0001) with both traits. These top 32 SNPs were all located on Chromosome BTA 14, between 21.95 Mb and 28.4 Mb. These results suggest that the genes located in that region of BTA 14 play a role in pubertal development in Brahman cattle. There are many annotated genes underlying this region of BTA 14 and these are the subject of current research. Further, we identified a region on Chromosome X where markers were associated (P < 1.00E–8) with AGE26, but not with AGECL. Information about specific genes and markers add value to our understanding of puberty and potentially contribute to genomic selection. Therefore, identifying these genes contributing to genetic variation in AGECL and AGE26 can assist with the selection for early onset of puberty.
Additional keywords: Bos indicus, corpus luteum, genome-wide association, scrotal circumference.
Introduction
Age at puberty is an important component of cattle performance. It determines the beginning of reproductive life for any breeding animal and it influences generation interval, affecting the rate of genetic improvement and herd productivity. Selecting for early pubertal heifers and bulls is a practical approach for reducing the generation interval and potentially increasing fertility (Lesmeister et al. 1973; Foster 1994; Siddiqui et al. 2008; Johnston et al. 2010).
For genetic selection, puberty must be defined as a measurable and inherited trait. Puberty in heifers was defined by plasma progesterone concentration, ultrasound images of corpus luteum (CL) and detection of oestrous (Shamay et al. 2005; Romano et al. 2007; Johnston et al. 2009). Puberty in bulls was defined by scrotal circumference thresholds and was related to sperm concentration, motility and morphology (Lunstra et al. 1978; Bagu et al. 2004; Siddiqui et al. 2008; Corbet et al. 2009). Pubertal traits vary in heritability (h2) from low to moderately high (Cammack et al. 2009).
There is a historical and increasing body of evidence reporting the genetic correlations between female and male puberty in cattle (Martin et al. 1992). Scrotal circumference of bulls correlates with puberty in their female relatives. Correlation estimates range from r = −0.71 to r = −0.15 (Burns et al. 2011). This evidence supports the indirect selection strategy for early pubertal heifers, using correlated traits measured in bulls. Given the size of the genetic correlation between these traits, some genes and genetic markers associated with puberty in heifers are likely to be associated with puberty in bulls, and vice versa.
Bos indicus cattle, such as Brahman cattle, are reportedly older at puberty when compared with most Bos taurus breeds (Lunstra and Cundiff 2003; Lopez et al. 2006). It is of economic relevance to select for early age at puberty in Brahman cattle. To assist selection and identify genes underlying fertility, we performed a GWAS in a sample of bulls with a measurement for puberty, namely the age at a scrotal circumference of 26 cm (AGE26). The present study is the first report of a GWAS regarding scrotal circumference in bulls, while the GWAS on AGECL in heifers represents a re-analysis of a previously reported study (Hawken et al. 2012). The Brahman heifers measured for AGECL were the mothers of the bulls measured for AGE26. Heritability, genetic correlation and genes associated with AGECL and AGE26 are reported and discussed.
Materials and methods
Cattle and traits
Cattle were bred and supplied by the Cooperative Research Centre for Beef Genetic Technologies (Beef CRC) as described previously (Corbet et al. 2009; Johnston et al. 2009). In brief, data from 1007 Brahman heifers and 1118 Brahman bulls were included in the current analysis. The phenotypes of the heifers have been reported previously (Johnston et al. 2009). The bulls included in the present experiment were the offspring of the heifers, which became breeding heifers for the Beef CRC. Their pedigree consists of over 50 grandsire families; 54 bulls that sired heifers, which were mated to 55 industry sires to produce the bulls used in the study.
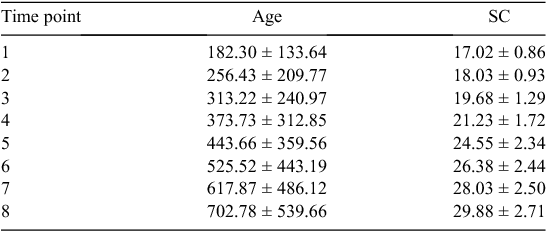
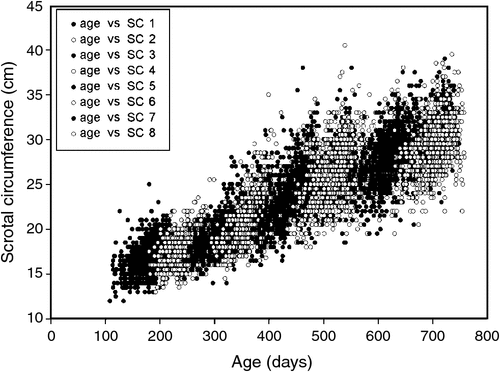
The traits that defined puberty were AGECL in heifers and AGE26 in bulls. The first CL was detected by ovarian ultrasound examinations conducted every 4–6 weeks, after heifers had reached an average liveweight of 200 kg (Johnston et al. 2009). AGECL was estimated from annotation of the date when the first CL was observed and the date of birth. Scrotal circumference (SC) was measured with a standard metal tape (Fordyce et al. 2006). Between weaning and 24 months of age, eight measurements of SC were taken for each bull, at 3-month intervals. Summary statistics for the age and SC of bulls at each of the eight time points are presented in Table 1. Using these repeated measurements for individual regressions, we interpolated the age when the bull achieved 26 cm of SC (AGE26, expressed in days). Achieving SC of 26 cm was considered a threshold for puberty in Bos indicus bulls. The threshold was different from the previously described 28-cm threshold because Bos indicus typically present a more elongated scrotum and a smaller SC than do Bos taurus (Lunstra et al. 1988; McGowan et al. 2002; Silva et al. 2011). For a visual assessment of age v. SC in this population, see Fig. 1. A more in-depth description and quantitative analysis of all reproductive and growth phenotypes available for this population of bulls was reported elsewhere (Corbet et al. 2009, 2011).
|
|
Genotypes
The Illumina BovineSNP50 array (Van Tassell et al. 2008; Matukumalli et al. 2009) was used to genotype samples according to the manufacturer’s protocols (Illumina Inc., San Diego, CA). Positions for each SNP were based on the UMD3 assembly of the bovine genome sequence (available from http://www.livestockgenomics.csiro.au/cgi-bin/gbrowse/btauUMD3/, verified 14 December 2011). Genotypes for 843 heifers had been generated previously using the BovineSNP50 v1 array and were reported in two recent GWAS that analysed growth traits and reproduction traits, including AGECL (Bolormaa et al. 2011; Hawken et al. 2012).
In the present study, 1118 bulls were genotyped using the Illumina BovineSNP50 v2 array. Quality control was similar to the previous study, with repeat samples included and the Bead Studio software from Illumina (www.illumina.com, verified 28 February 2012) used to determine the genotype calls. Animals with call rates <98% were excluded, resulting in 1105 bulls being retained for analyses. SNPs with auto-calling rates <85% and SNPs with a minor allele frequency <0.01 were excluded from further analyses.
For the current study, missing genotypes for bulls and heifers were imputed using the BEAGLE 3.2 program (Browning and Browning 2010). Imputation and quality-control procedures yielded a total of 43 821 SNPs used in the genome-wide association analysis. Genotype calls were coded as 0 for the homozygote of the first allele (A), 1 for the heterozygote, and 2 for the homozygote of the second allele (B). Alleles A and B were defined as per top and bottom rules from Illumina (http://www.illumina.com/documents/products/technotes/technote_topbot.pdf, verified 17 February 2012). Imputation resulted in better coverage of the X chromosome, adding new data to the previous heifer study on AGECL (Hawken et al. 2012). This improved X chromosome coverage for the heifers and all genotypes obtained for bulls are reported here for the first time.
Genetic correlation and heritability
Genetic correlation and h2 (for AGE26) were estimated fitting the two traits in a bivariate analysis using a mixed model and three generation pedigree relationships. For AGECL, fixed effects included in the model and h2 have been reported previously (Johnston et al. 2009); here, we estimated the h2 de novo in the bivariate analysis, using the same fixed effects. Fixed effects included in the model for AGE26 were contemporary group (bulls born in the same year and location – or cohort – and post-weaning location) and birth month. Solutions to the effects in the model as well as variance components were estimated using VCE v.6 software (Groeneveld and García-Cortés; http://vce.tzv.fal.de, verified 20 February 2012).
Genome-wide association study
Genome-wide association studies (GWAS) were performed for AGECL and AGE26 separately and by using two approaches. First, GWAS was performed with the full datasets for bulls (1105) and heifers (843) and, subsequently, each population was split into calibration and a validation datasets, for providing additional evidence of SNP association and estimated effects. The calibration and validation datasets were generated according to birth date, dividing the populations into a younger half (~50%) and an older half (~50%). For AGECL, the calibration dataset was formed by heifers born until November 2001 (n = 480) and older half was born from December 2001 onwards (n = 363). For AGE26, the calibration dataset comprised those bulls born until December 2005 (n = 565) and the bulls in the validation dataset were born from January 2006 onwards (n = 550). Using birth date to define calibration and validation datasets is a tested approach (Hayes et al. 2010). A GWAS for AGECL using the full dataset has been reported previously (Hawken et al. 2012), but the results reserving 363 heifers for a validation exercise are reported only here. No previous study exists to relate the GWAS for AGE26 reported here.
The effect of each SNP was estimated three times, as follows: first, using phenotypic data from the full dataset; second, using phenotypic data from the calibration dataset; and finally, using phenotypic data from the validation dataset. In all three instances, the effect of each SNP was estimated using the mixed model of Eqn 1, as follows:

where yij represents the vector of observations from the ith heifer or bull at the jth trait (j = 1 and 2 for AGECL and AGE26, respectively); X is the incidence matrix relating fixed effects in β with observations in yij; Z is the incidence matrix relating random additive polygenic effects in u with observations in yij; Sk in the vector of genotypes for the kth SNP across all animals; sjk represents the additive association of the kth SNP on the jth trait; and eij is the vector of random residual effects. Fixed effects included in the model were the same as described for the models used to estimate the genetic correlations and h2. Tests for the significance of SNP association were conducted using Qxpak5 (Perez-Enciso and Misztal 2004, 2011). Qxpak5 performs a likelihood ratio test, testing the model with SNP versus the model without the SNP, against a chi-square distribution with 1 degree of freedom and this was carried out one SNP at a time. Chromosome X genotypes of males and females were not analysed together, because the traits were analysed separately.
False discovery rates were estimated using Eqn 2, as follows:

where n represents the total number of SNPs included in the study (in the present study, n = 43 821), P is the P-value threshold being used and k is the actual number of associated SNPs in the given P-value threshold.
The percentage of the genetic variance accounted for by the ith SNP was computed according to the formulae of Eqn 3, as follows:

where pi and qi are the allele frequencies for the ith SNP estimated across the entire population, ai is the estimated additive effect of the ith SNP on the trait in question (AGECL or AGE26), and σg2 is the REML estimate of the (poly-)genetic variance for the trait in question.
Results
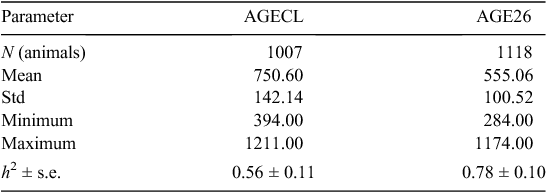
Descriptive statistics, h2 and associated errors for AGECL and AGE26 are presented in Table 2. At puberty, heifers were on average older (751 ± 142 days) than bulls (555 ± 101 days). The heritability estimated for puberty in heifers was lower (AGECL, h2 = 0.56 ± 0.11) than that in bulls (AGE26, h2 = 0.78 ± 0.10). The traits AGECL and AGE26 were genetically correlated, with r = 0. 20 ± 0.10 estimated using REML methods.
|
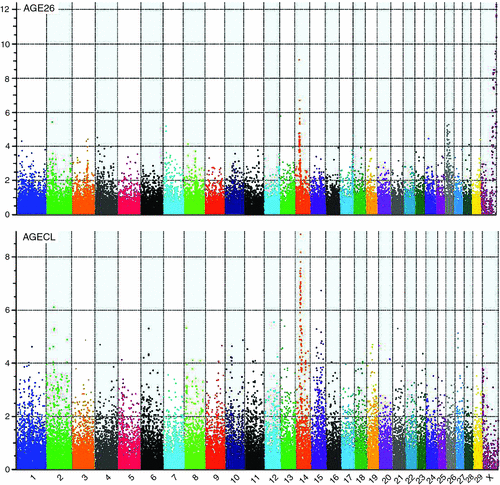
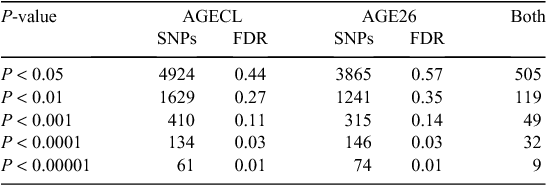
Figure 2 provides an overview of GWAS results for the full datasets presented as Manhattan plots (i.e. significance on the y-axis v. genome map position on the x-axis) for AGE26 and AGECL. Two regions showed clear association peaks in the GWAS of AGE26, one on the X chromosome and another on BTA14. The GWAS of AGECL also showed a peak on BTA14 for the same region where SNPs were significant for AGE26. There were 134 SNPs associated with AGECL and 146 SNPs associated with AGE26 (P < 0.0001, false discovery rate = 0.03). Of these SNPs, 32 (~22%) were associated with both traits (P < 0.0001, Table 3). These 32 SNPs that were in common for both traits were all located on BTA14, between 21.95 Mb and 28.4 Mb. The most significant SNP for AGECL was Hapmap23509-BTC-073113 at Position 27 198 715 of BTA14 (P = 1.36E–09). The most significant SNP for AGE26 was BTA-30242-no-rs at Position 85 495 447 of Chromosome X (P = 5.35E–13). The effects, P-values and proportion of genetic variance explained for each of the 32 SNPs associated (P < 0.0001) with both AGECL and AGE26 are presented in Table 4. The effects, P-values and proportion of the variance explained for the SNPs underlying the association peak located on X chromosome are also reported (Table 5).
|
|
The nearest annotated genes and their distance to the reported SNPs were determined using the UMD3.0 bovine genome assembly (Tables 4, 5). However, to consider only the genes nearest to each SNP could be misleading. It is preferable to consider all genes mapped to the association regions. In total, 23 genes mapped to the association region in Chromosome 14, including ATP6V1H, CA8, CHD7, CSF2RA, FAM110B, IMPAD1, NPBWR1, NSMAF, OPRK1, PCMTD1, PENK, POLR2K, RAB2A, RB1CC1, RGS20, RLBP1 L1, RP1, SDR16C6, SNTG1, SOX17, TGS1, TOX and XKR4. Also, 46 genes mapped to the association region in Chromosome X, including ABCB7, AKAP4, APOL, AR, ARR3, BRWD3, CACNA1F, CCDC120, CHIC1, CHM, CLCN5, CXCR3, CYLC1, DGAT2 L3, DGAT2 L6, EDA, EFNB1, FGF16, FOXP3, GPKOW, H11CXORF26, KIF4A, LRCH2, MGC140080, MGC152340, MIR374A, MIR374B, NHSL2, PAGE4, PJA1, PLS3, POU3F4, RBM10, SLC9A7, SNX12, STARD8, SUV39H1, TAF1, TAF9B, TIMM17B, TMEM28, UPRT, USP11, USP27X, ZDHHC15 and ZNF182.
Manhattan plots for the calibration and validation datasets are provided in the Supplementary Material Fig. S1. There were 111 SNPs that were associated (P < 0.01) with AGE26 in both calibration and validation datasets (Supplementary Material Table S1). There were 98 SNPs that were associated (P < 0.01) with AGECL in both calibration and validation datasets (Supplementary Material Table S2). Chromosome X harboured more SNPs associated with AGE26, in both calibration and validation sets, than did any other chromosome (49%, or 55 of 111 SNPs), followed by BTA 14 (20%, or 23 of 111 SNPs). Most (58%, or 57 of 98 SNPs) of the SNPs associated with AGECL in both calibration and validation sets were located at BTA 14. As a result of our calibration and validation exercise, the same regions and candidate genes as observed in the GWAS with the full dataset remained relevant.
Discussion
Heritability estimates of cattle puberty in the literature are variable and influenced by population, breed, management and environmental factors, as well as the use of different phenotypic measurements (Martin et al. 1992; Cammack et al. 2009). Heritability for age at puberty in the Beef CRC Brahman heifers (AGECL, h2 = 0.57) has been reported previously (Johnston et al. 2009); we confirmed these results by estimating a very similar value (h2 = 0.56). These heritabilities are within the range, from 0.20 to 0.67, of other estimates for age at puberty (Arije and Wiltbank 1971; Smith et al. 1976; Laster et al. 1979). Reported heritability estimates for scrotal circumference range from 0.39 to 0.75 (Lunstra et al. 1988; Martinez-Velazquez et al. 2003; Corbet et al. 2009). Our result for puberty in bulls (AGE26) was closer to the higher end of that range (h2 = 0.78). Overall and in agreement with our results, the heritability of puberty in cattle ranges from moderate to high, allowing for improvement through genetic selection.
Pubertal traits measured in heifers (AGECL) and bulls (AGE26) had a positive and favourable genetic correlation. This result confirms previous reports that showed that bigger scrotal circumference in bulls was correlated with early puberty in their female relatives (Martin et al. 1992; Burns et al. 2011). Previous evidence and our results point to the practicality of selecting for early pubertal cattle because both heifers and bulls can be selected with beneficial correlated effects.
Considering the genetic correlation, genes and SNPs associated with puberty in heifers were likely to be relevant for puberty in bulls, and vice versa. On BTA14, a large number of SNPs associated with puberty were identified in both bulls and heifers. The SNPs on BTA14 are located close to RP1, XKR4 and TOX. These genes are not known to affect puberty in any species. However, the region on BTA14 includes another 20 annotated genes and presents homology to Human chromosome HSA8q12, which is associated with height (Gudbjartsson et al. 2008). Among those 20 annotated genes, PENK, RPS20, SNORD54 and PLAG1 are plausible functional candidates. PENK has a role in GnRH regulation (Rosie et al. 1992; Taylor et al. 2007). RPS20 and a small RNA (SNORD54) were recently associated with calving ease (Pausch et al. 2011). The alleles that lower calving-ease have a positive effect on growth traits (Pausch et al. 2011). Calving ease is a trait influenced by the size of the calf and so, similarly to stature, it reflects frame size. Cattle with smaller frame size achieve puberty earlier than those with larger frame size (Vargas et al. 1999). Taken together, this evidence could point to RPS20 and SNORD54 as pleiotropic genes affecting puberty and calving-ease mainly through their effect on growth. Another recent study argued that PLAG1 was the relevant gene underlying this region on BTA14 and affecting bovine stature (Karim et al. 2011). Considering the physiological link between growth and puberty, it is possible to hypothesise that the association region in BTA14 may relate to a gene with many pleiotropic effects or a functional polymorphism that affects more than one gene. There is already evidence for a functional SNP in this region affecting the expression levels of multiple genes (Karim et al. 2011).
The two most significant SNPs for AGE26 were located on the X chromosome and not on BTA14. The genes nearest to the most significant SNPs, EDA and DGAT2 L3, do not appear to play a role in reproduction, given the current evidence from literature. A formal discussion of the functional link between these genes and cattle puberty is beyond our present objective. However, the association peak on Chromosome X does include the following three genes that are positional and functional candidate genes: AR (androgen receptor), TAF1 and TAF9B (both TATA box binding protein (TBP)-associated factors). These genes are candidate genes for several reasons. The androgen receptor is known to play a role in sexual development, specifically, it affects spermatogenesis, testis localisation and testis size in mice models (Verhoeven et al. 2010). In pigs, the AR is considered a candidate gene for reproduction and performance traits (Trakooljul et al. 2004). Manipulation of fetal androgen exposure alters the timing of puberty in sheep (Jackson et al. 2008). The genes TAF1 and TAF9B encode transcription factors that form the TFIID complex, a regulator of cell cycle and differentiation. Testis-specific TAF proteins have been reported as relevant for spermatid differentiation in Drosophila (Hiller et al. 2004). Taken together, this evidence and our results support AR, TAF1 and TAF9B as candidate genes for scrotal development and puberty in Brahman bulls.
Results of the present study are consistent with the hypothesis that the genes underlying the associated regions on BTA14 and Chromosome X play significant roles on defining age at puberty in Brahman cattle. Candidate genes mapping to these regions are the subject of ongoing research. Information about specific genes and markers will add value to genomic selection. Therefore, identifying these associated regions contributing to genetic variation in AGECL and AGE26 should assist in the selection for early onset of puberty in Brahman cattle.
Acknowledgements
Data reported herein were generated as part of a larger study performed by the Cooperative Research Centre for Beef Genetic Technologies. Meat and Livestock Australia, Department of Employment, Economic Development and Innovation (DEEDI, Queensland government), CSIRO Livestock Industries, Australian Centre for International Agricultural Research, Northern Pastoral Group and University of Queensland supported the project. We gratefully acknowledge those who measured traits and managed cattle or DNA samples, including Richard Holroyd, Matthew Lee Wolcott, Yuandan Zhang, Mick Sullivan, Tim Grant, Brian Burns, Neil Cooper, Peggy Olsson, Debrah Corbet, Bronwyn Venus, Tracy Longhurst, Paul Williams, Warren Sim, Rob Young, Laercio R. Porto Neto, Rowan Bunch, Russell McCulloch, Blair Harrison, Eliza Collis, Mike Goddard, Brett Mason, Keith Savin and Kathryn Guthridge.
References
Arije GF, Wiltbank JN (1971) Age and weight at puberty in Hereford heifers. Journal of Animal Science 33, 401–406.Bagu ET, Madgwick S, Duggavathi R, Bartlewski PM, Barrett DM, Huchkowsky S, Cook SJ, Rawlings NC (2004) Effects of treatment with LH or FSH from 4 to 8 weeks of age on the attainment of puberty in bull calves. Theriogenology 62, 861–873.
| Effects of treatment with LH or FSH from 4 to 8 weeks of age on the attainment of puberty in bull calves.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXls1aisbY%3D&md5=b8dff51a76137c283797eda0e7570666CAS |
Bolormaa S, Hayes BJ, Savin K, Hawken R, Barendse W, Arthur PF, Herd RM, Goddard ME (2011) Genome-wide association studies for feedlot and growth traits in cattle. Journal of Animal Science 89, 1684–1697.
| Genome-wide association studies for feedlot and growth traits in cattle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXnsVWktr0%3D&md5=2d87660a340ad5b0c8374e656685e5e4CAS |
Browning SR, Browning BL (2010) High-resolution detection of identity by descent in unrelated individuals. American Journal of Human Genetics 86, 526–539.
| High-resolution detection of identity by descent in unrelated individuals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXlsFWmsb8%3D&md5=9811e9750bbc16ffcf5fe37a83076c93CAS |
Burns B, Gazzola C, Holroyd R, Crisp J, McGowan M (2011) Male reproductive traits and their relationship to reproductive traits in their female progeny: a systematic review. Reproduction in Domestic Animals 46, 534–553.
| Male reproductive traits and their relationship to reproductive traits in their female progeny: a systematic review.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3MvoslWiug%3D%3D&md5=d0beb61611a4bc9582306f19adc8627aCAS |
Cammack KM, Thomas MG, Enns RM (2009) Reproductive traits and their heritabilities in beef cattle. The Professional Animal Scientist 25, 517–528.
Corbet NJ, Burns BM, Corbet DH, Johnston DJ, Crisp JM, McGowan M, Prayaga KC, Venus BK, Holroyd RG (2009) Genetic variation in groth, hormonal and seminal traits of young tropically adapted bulls. Proceedings of the Association for the Advancement of Animal Breeding and Genetics 18, 121–124.
Corbet NJ, Burns B, Corbet DH, Crisp JM, Johnston DJ, McGowan MR, Venus BK, Holroyd RG (2011) Bull traits measured early in life as indicators of herd fertility. Proceedings of the Association for the Advancement of Animal Breeding and Genetics 19, 55–58.
Fordyce G, Entwistle K, Norman S, Perry V, Gardiner B, Fordyce P (2006) Standardising bull breeding soundness evaluations and reporting in Australia. Theriogenology 66, 1140–1148.
| Standardising bull breeding soundness evaluations and reporting in Australia.Crossref | GoogleScholarGoogle Scholar |
Foster DL (1994) Puberty in the sheep. In ‘The Physiology of Reproduction’. 2nd edn. (Eds E Knobil, JD Neill). (Raven Press Ltd: New York)
Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Pedersen O, Aben KK, Witjes JA, Swinkels DW, den Heijer M, Franke B, Verbeek ALM, Becker DM, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Gulcher J, Kiemeney LA, Kong A, Thorsteinsdottir U, Stefansson K (2008) Many sequence variants affecting diversity of adult human height. Nature Genetics 40, 609–615.
| Many sequence variants affecting diversity of adult human height.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXltFylu74%3D&md5=a1e358412c3c1f7b9b17cfe0c6a890d9CAS |
Hawken RJ, Zhang YD, Fortes MRS, Collis E, Barris WC, Corbet NJ, Williams PJ, Fordyce G, Holroyd RG, Walkley JRW, Barendse W, Johnston DJ, Prayaga KC, Tier B, Reverter A, Lehnert SA (2012) Genome-wide association studies of female reproduction in tropically adapted beef cattle. Journal of Animal Science
| Genome-wide association studies of female reproduction in tropically adapted beef cattle.Crossref | GoogleScholarGoogle Scholar | in press.
Hayes BJ, Pryce J, Chamberlain AJ, Bowman PJ, Goddard ME (2010) Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLOS Genetics 6, e1001139
| Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits.Crossref | GoogleScholarGoogle Scholar |
Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT (2004) Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131, 5297–5308.
| Testis-specific TAF homologs collaborate to control a tissue-specific transcription program.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVCjs7bO&md5=f969b808001499b97e0dcedbaa9174feCAS |
Jackson LM, Timmer KM, Foster DL (2008) Sexual differentiation of the external genitalia and the timing of puberty in the presence of an antiandrogen in sheep. Endocrinology 149, 4200–4208.
| Sexual differentiation of the external genitalia and the timing of puberty in the presence of an antiandrogen in sheep.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXptlamtb8%3D&md5=bfb0d1f25ce696a69aaa58fc36f28dddCAS |
Johnston DJ, Barwick SA, Corbet NJ, Fordyce G, Holroyd RG, Williams PJ, Burrow HM (2009) Genetics of heifer puberty in two tropical beef genotypes in northern Australia and associations with heifer- and steer-production traits. Animal Production Science 49, 399–412.
| Genetics of heifer puberty in two tropical beef genotypes in northern Australia and associations with heifer- and steer-production traits.Crossref | GoogleScholarGoogle Scholar |
Johnston D, Barwick S, Fordyce G, Holroyd R (2010) Understanding the genetics of lactation anoestrus in Brahman beef cattle to enhance genetic evaluation of female reproductive traits. In ‘9th world congress on genetics applied to livestock production’. (German Society for Animal Science: Leipzig, Germany)
Karim L, Takeda H, Lin L, Druet T, Arias JAC, Baurain D, Cambisano N, Davis SR, Farnir F, Grisart B, Harris BL, Keehan MD, Littlejohn MD, Spelman RJ, Georges M, Coppieters W (2011) Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nature Genetics 43, 405–413.
| Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXltFyqsrY%3D&md5=db9b3885d5ab2fd5f2344016bb66d07dCAS |
Laster DB, Smith GM, Cundiff LV, Gregory KE (1979) Characterization of biological types of cattle (Cycle-II). 2. Post-weaning growth and puberty of heifers. Journal of Animal Science 48, 500–508.
Lesmeister JL, Burfening PJ, Blackwell RL (1973) Date of first calving in beef cows and subsequent calf production. Journal of Animal Science 36, 1–6.
Lopez R, Thomas MG, Hallford DM, Keisler DH, Silver GA, Obeidat BS, Garcia MD, Krehbiel CR (2006) Case study: metabolic hormone profiles and evaluation of associations of metabolic hormones with body fat and reproductive characteristics of Angus, Brangus and Brahman heifers. The Professional Animal Scientist 22, 273–282.
Lunstra DD, Cundiff LV (2003) Growth and pubertal development in Brahman-, Boran-, Tuli-, Belgian Blue-, Hereford- and Angus-sired F1 bulls. Journal of Animal Science 81, 1414–1426.
Lunstra DD, Ford JJ, Echternkamp SE (1978) Puberty in beef bulls: hormone concentrations, growth, testicular development, sperm production and sexual aggressiveness in bulls of different breeds. Journal of Animal Science 46, 1054–1062.
Lunstra DD, Gregory KE, Cundiff LV (1988) Heritability estimates and adjustment factors for the effects of bull age and age of dam on yearling testicular size in breeds of bulls. Theriogenology 30, 127–136.
| Heritability estimates and adjustment factors for the effects of bull age and age of dam on yearling testicular size in breeds of bulls.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD283pvFahsA%3D%3D&md5=0e12e46ff42be1c64c225ccb7b166da2CAS |
Martin LC, Brinks JS, Bourdon RM, Cundiff LV (1992) Genetic effects on beef heifer puberty and subsequent reproduction. Journal of Animal Science 70, 4008–4017.
Martinez-Velazquez G, Gregory KE, Bennett GL, Van Vleck LD (2003) Genetic relationships between scrotal circumference and female reproductive traits. Journal of Animal Science 81, 395–401.
Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP, O’Connell J, Moore SS, Smith TPL, Sonstegard TS, Van Tassell CP (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 4, e5350
| Development and characterization of a high density SNP genotyping assay for cattle.Crossref | GoogleScholarGoogle Scholar |
McGowan MR, Bertram JD, Fordyce G, Fitzpatrick LA, Miller RG, Jayawardhana GA, Doogan VJ, De Faveri J, Holroyd RG (2002) Bull selection and use in northern Australia. 1. Physical traits. Animal Reproduction Science 71, 25–37.
| Bull selection and use in northern Australia. 1. Physical traits.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD383kvFSrsA%3D%3D&md5=92d5cfbb7680dcc70924dd3460baf882CAS |
Pausch H, Flisikowski K, Jung S, Emmerling R, Edel C, Gotz KU, Fries R (2011) Genome-wide association study identifies two major loci affecting calving ease and growth-related traits in cattle. Genetics 187, 289–297.
| Genome-wide association study identifies two major loci affecting calving ease and growth-related traits in cattle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXisVyiurY%3D&md5=e275ad5ecc39e695a0e38b9e07e27386CAS |
Perez-Enciso M, Misztal I (2004) Qxpak: a versatile mixed model application for genetical genomics and QTL analyses. Bioinformatics 20, 2792–2798.
| Qxpak: a versatile mixed model application for genetical genomics and QTL analyses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVSru7zN&md5=1a1a3ad3f2e3358de76aa8b069dce622CAS |
Perez-Enciso M, Misztal I (2011) Qxpak.5: Old mixed model solutions for new genomics problems. BMC Bioinformatics 12, 202
| Qxpak.5: Old mixed model solutions for new genomics problems.Crossref | GoogleScholarGoogle Scholar |
Romano MA, Barnabe VH, Kastelic JP, de Oliveira CA, Romano RM (2007) Follicular dynamics in heifers during pre-pubertal and pubertal period kept under two levels of dietary energy intake. Reproduction in Domestic Animals 42, 616–622.
| Follicular dynamics in heifers during pre-pubertal and pubertal period kept under two levels of dietary energy intake.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD2snltFGlsA%3D%3D&md5=18cf78caca522722b846b42c3128e82cCAS |
Rosie R, Thomson E, Blum M, Roberts JL, Fink G (1992) Oestrogen positive feedback reduces arcuate proopiomelanocortin messenger ribonucleic acid. Journal of Neuroendocrinology 4, 625–630.
| Oestrogen positive feedback reduces arcuate proopiomelanocortin messenger ribonucleic acid.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXhs1Cktg%3D%3D&md5=4f68f98ecd7983fdddbf0a74d40a0d75CAS |
Shamay A, Werner D, Moallem U, Barash H, Bruckental I (2005) Effect of nursing management and skeletal size at weaning on puberty, skeletal growth rate, and milk production during first lactation of dairy heifers. Journal of Dairy Science 88, 1460–1469.
| Effect of nursing management and skeletal size at weaning on puberty, skeletal growth rate, and milk production during first lactation of dairy heifers.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXivVykuro%3D&md5=8c53aa53df548a6caa1363176519f0c6CAS |
Siddiqui MA, Bhattacharjee J, Das ZC, Islam MM, Islam MA, Haque MA, Parrish JJ, Shamsuddin M (2008) Crossbred bull selection for bigger scrotum and shorter age at puberty with potentials for better quality semen. Reproduction in Domestic Animals 43, 74–79.
Silva MR, Pedrosa VB, Silva JCB, Eler JP, Guimarães JD, Albuquerque LG (2011) Testicular traits as selection criteria for young Nellore bulls. Journal of Animal Science 89, 2061–2067.
| Testicular traits as selection criteria for young Nellore bulls.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXoslCis7w%3D&md5=664667665a7c6b34d1b4c0268c92aa51CAS |
Smith GM, Fitzhugh HA, Cundiff LV, Cartwright TC, Gregory KE (1976) Genetic-analysis of maturing patterns in straightbred and crossbred Hereford, Angus and Shorthorn cattle. Journal of Animal Science 43, 389–395.
Taylor JA, Goubillon ML, Broad KD, Robinson JE (2007) Steroid control of gonadotropin-releasing hormone secretion: associated changes in pro-opiomelanocortin and preproenkephalin messenger RNA expression in the ovine hypothalamus. Biology of Reproduction 76, 524–531.
| Steroid control of gonadotropin-releasing hormone secretion: associated changes in pro-opiomelanocortin and preproenkephalin messenger RNA expression in the ovine hypothalamus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXitlWhu7k%3D&md5=525e5c8b0db100c3cf1b222d0f8d2ac3CAS |
Trakooljul N, Ponsuksili S, Schellander K, Wimmers K (2004) Polymorphisms of the porcine androgen receptor gene affecting its amino acid sequence and expression level. Biochimica et Biophysica Acta (BBA) – Gene Structure and Expression 1678, 94–101.
| Polymorphisms of the porcine androgen receptor gene affecting its amino acid sequence and expression level.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXkt1Smtb4%3D&md5=e3f88a1d9b4497cc39c3910c2c9c3f5aCAS |
Van Tassell CP, Smith TP, Matukumalli LK, Taylor JF, Schnabel RD, Lawley CT, Haudenschild CD, Moore SS, Warren WC, Sonstegard TS (2008) SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nature Methods 5, 247–252.
| SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXisFOmtLk%3D&md5=f586ca732d48c9fb462a03fc3fe3eef2CAS |
Vargas CA, Olson TA, Chase CC, Hammond AC, Elzo MA (1999) Influence of frame size and body condition score on performance of Brahman cattle. Journal of Animal Science 77, 3140–3149.
Verhoeven G, Willems A, Denolet E, Swinnen JV, De Gendt K (2010) Androgens and spermatogenesis: lessons from transgenic mouse models. Philosophical Transactions of the Royal Society B Biological Sciences 365, 1537–1556.
| Androgens and spermatogenesis: lessons from transgenic mouse models.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtVaisr3L&md5=7d90bb446f36e7692bd9b7fa7cb3ac48CAS |




