Molecular evidence for association of Tomato leaf curl New Delhi virus with leaf curl disease of papaya (Carica papaya L.) in India
S. K. Raj A C , S. K. Snehi A , M. S. Khan A , R. Singh A and A. A. Khan BA Molecular Virology Laboratory, National Botanical Research Institute, Lucknow, UP 226001, India.
B Plant Virology Laboratory, Aligarh Muslim University, Aligarh, UP 202002, India.
C Corresponding author. Email: skraj2@rediffmail.com
Australasian Plant Disease Notes 3(1) 152-155 https://doi.org/10.1071/DN08059
Submitted: 18 May 2008 Accepted: 10 November 2008 Published: 20 November 2008
Abstract
Association of Tomato leaf curl New Delhi virus with leaf curl disease of papaya (Carica papaya L.) was detected by polymerase chain reaction using begomovirus-specific primers and confirmed by highest sequence similarities and close phylogenetic relationships.
Papaya (Carica papaya L.) is cultivated commercially throughout the world’s tropical and subtropical regions for its edible fruits. Papain, an enzyme prepared from dried latex of immature fruits, is used for meat tenderising, food processing and in the leather industry (Singh 2006). The vasculature latex of papaya has medicinal value and is used to treat ulcers, dissolve membranes in diphtheria, and reduce swelling, fever and adhesions after surgery (Singh et al. 1983). The limiting factor of papaya cultivation is its susceptibility to ring spot, leaf curl, mosaic and distortion diseases. Among them leaf curl disease caused by Begomovirus species is one of the most serious threats to papaya cultivation in most of the papaya growing countries (Singh 2006). Various RNA viruses have been reported on papaya worldwide, viz. Papaya ring spot virus (Potyvirus), Papaya mosaic virus (Potexvirus), Tomato spotted wilt virus (Tospovirus), Papaya apical necrosis virus (Rhabdovirus), Tobacco ring spot virus (Nepovirus), Tobacco streak virus (Ilarvirus), Tobacco rattle virus (Tobravirus) and Cucumber mosaic virus (Cucumovirus) (Singh 2006). However, a limited number of DNA viruses are known to be associated with the leaf curl disease of papaya: Papaya leaf curl virus (Nadeem et al. 1997; Singh 2006), Papaya leaf curl China virus and Papaya leaf curl Guangdong virus (Wang et al. 2004), and Papaya leaf curl Taiwan virus (Chang et al. 2003). These viruses are members of the Begomovirus group of the family Geminiviridae. The family comprises viruses with circular single stranded DNA genomes encapsidated in geminate quasi-isometric virion particles of ~20–30 nm in size which are transmitted through whitefly (Bemisia tabaci) (Harrison 1985).
A typical leaf curl disease was observed on papaya grown in and around Lucknow (India) during November 2006. Naturally infected papaya plants showed severe downward leaf curling, swelling of veins, twisting and reduction of petioles, and stunted growth of the whole plant which bore a few small, distorted fruit (Fig. 1a, b) compared with the healthy one (Fig. 1c). The whitefly population was also noticed in the papaya growing area, therefore, transmission of the disease was attempted through whiteflies (B. tabaci) in an insect proof glasshouse using an acquisition access period (AAP) of 18 h and inoculation access period (IAP) of 24 h. The disease was successfully transmitted by B. tabaci from naturally infected papaya to healthy seedlings of papaya, tobacco, tomato and chilli and all the inoculated test species displayed typical leaf curl symptoms indicating the association of a whitefly transmissible infectious agent, possibly a begomovirus, with the leaf curl disease of papaya.

|
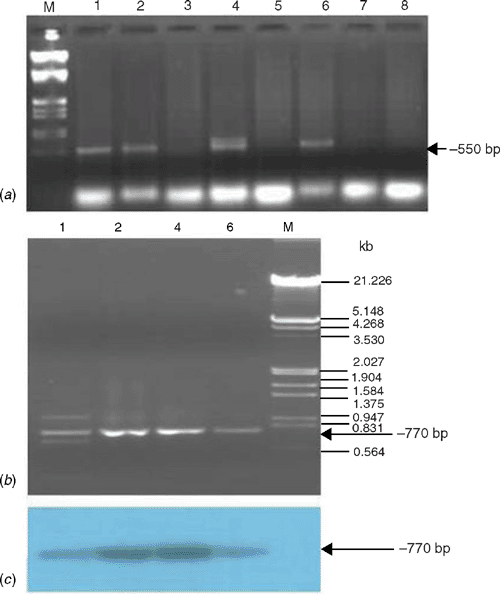
For molecular detection, the total DNA was extracted from 100 mg leaf tissues of naturally infected and healthy papaya samples collected from six locations by a method described earlier (Dellaporta et al. 1983) and subjected to polymerase chain reaction (PCR). The PCR was performed using begomovirus genus specific degenerate primers (Wyatt and Brown 1996) and begomovirus coat protein (CP) region specific primers (Singh 2005) in a 50 μL reaction mixture containing: template DNA (100 ng), dNTPs (10 mmol each), primers (each 25 pmol), Pfu DNA polymerase (1.5 U), assay buffer (10×) in an automated thermal cycler (MJ Research, USA). The PCR conditions were: initial denaturation at 94°C for 5 min followed by 30 cycles of PCR of 1 min at 94°C, 30 s at 52°C (for Wyatt and Brown 1996 primers) or 47°C (for Singh 2005 primers) and extension at 72°C for 1.5 min, and a final extension of 5 min at 72°C. The PCR products were analysed by electrophoresis in 1.2% agarose gels. As expected, a band of ~550 bp in 4 of 6 symptomatic samples was successfully amplified by the Wyatt and Brown (1996) primers; however, no such amplicon was obtained in 2 of 2 healthy samples collected from the same location (Fig. 2a). The Singh (2005) primers were then used for PCR on DNA that had previously tested positive by PCR using the Wyatt and Brown (1996) primers and these all gave ~770 bp band (Fig. 2b). To confirm the authenticity of the ~770 bp amplicons, the gel was blotted onto nylon membrane and was allowed to hybridise with a probe of a CP clone of Tomato leaf curl New Delhi virus (DQ431846) which showed strong signals of hybridisation with the probe (Fig. 2c), indicating that PCR amplicons originated from the CP region of the begomovirus. Presence of begomovirus was also confirmed by PCR by begomovirus CP-specific primers in test plants that had been infected through transmission by whitefly.
|
The ~770 bp amplicons obtained from three leaf samples were cloned into the pGEM-T easy vector system-1 (Promega Life Corporation, USA). One clone of each sample was sequenced in both orientations using SP6 and T7 primers (Bangalore Genei Pvt. Ltd, India). The consensus sequence data of three identical sequences, producing a complete CP gene sequence of 771 nucleotides, encoding 256 amino acid residues, was deposited in the GenBank database (Accession EF194275). Basic Local Alignment Search Tool (BLAST) analysis of the papaya isolate (EF194275) revealed highest 95–97% nucleotide sequence identities with isolates of Tomato leaf curl New Delhi virus (ToLCNDV, AY691902, AY691899, AM286433, AY939926 and EU366163).
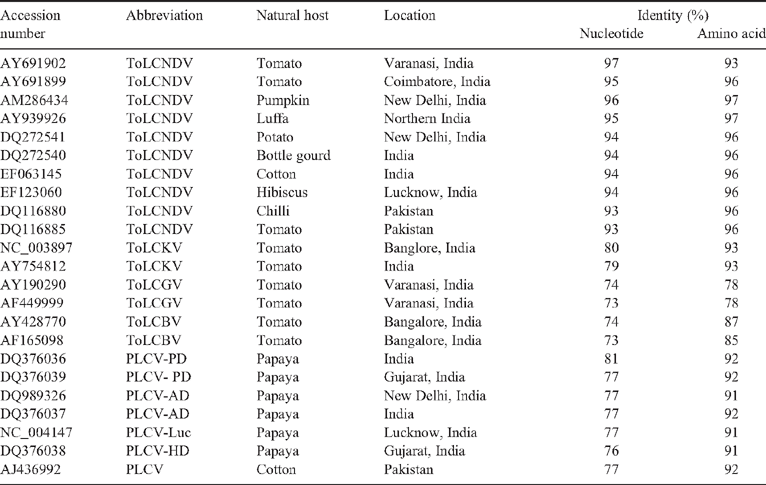
The Genomatix DiAlign2 program (Morgenstern 1998) was used to align the new begomovirus nucleotide and amino acid sequences (EF194275) with selected begomoviruses from a diverse range of host species including tomato, luffa, cotton, hibiscus, bottle gourd, and chilli (Table 1). EF194275 showed maximum 93–97% identities (at both nucleotide and amino acid levels) with ToLCNDV isolates. The identities of the new isolate with Papaya leaf curl virus (PLCV) isolates PLCV-PD, PLCV-AD, PLCV-HD, PLCV-Luc were low: 76–81% and 91–92% at nucleotide and amino acid levels, respectively (Table 1).
On the basis of the positive whitefly transmission test, PCR amplification of fragments of the expected size (~550 bp with begomovirus genus specific degenerate primers or ~770 bp with begomovirus CP specific primers), highest (96%) sequence identities with isolates of ToLCNDV and in accordance with the latest report of the International Committee on Taxonomy of Viruses (ICTV, Fauquet et al. 2008), the virus associated with leaf curl disease of papaya was identified as an isolate of ToLCNDV and designated as ToLCNDV-Papaya.
Papaya leaf curl disease has been reported from India by Thomas and Krishanaswamy (1939) and by Nariani (1956). The disease was known to be transmitted by the whitefly (B. tabaci) in a persistent manner and a begomovirus was detected from papaya by nucleic acid hybridisation tests (Saxena et al. 1998a, 1998b). Papaya leaf curl virus (PLCV) has been reported from Pakistan (Nadeem et al. 1997), Taiwan (Chang et al. 2003), China (Wang et al. 2004; Zhang et al. 2005; Huang and Zhou 2006) and India (Singh 2006). To the best of our knowledge this is a first report on association of ToLCNDV with leaf curl disease of papaya in India.
Acknowledgements
The authors express their gratitude to the Director, National Botanical Research Institute, Lucknow for facilities and Department of Science and Technology, New Delhi for funding to AA Khan.
Chang LS,
Lee YS,
Su HJ, Hung TH
(2003) First report of Papaya leaf curl virus Infecting papaya plants in Taiwan. Plant Disease 87, 204.
| Crossref | GoogleScholarGoogle Scholar |

Dellaporta SJ,
Wood J, Hick JB
(1983) A plant DNA minipreparation: Version II. Plant Molecular Biology Reporter 1, 19–21.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Fauquet CM,
Briddon RW,
Brown JK,
Moriones E,
Stanley J,
Zerbini M, Zhou X
(2008) Geminivirus strain demarcation and nomenclature. Archives of Virology 153, 783–821.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Harrison BD
(1985) Advances in Geminivirus research. Annual Review of Phytopathology 23, 55–82.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Huang JF, Zhou XP
(2006) First report of Papaya leaf curl China virus infecting Corchoropsis timentosa in China. Plant Pathology 55, 291.
| Crossref | GoogleScholarGoogle Scholar |

Morgenstern B
(1998) DiAlign2 Improvement of the segment to the segment approach to multiple sequence alignment. Bioinformatics (Oxford, England) 15, 203–210.

Nadeem A,
Mehmood T,
Tahir M,
Khalid S, Xing Z
(1997) First report of papaya leaf curl disease in Pakistan. Plant Disease 81, 1333.
| Crossref | GoogleScholarGoogle Scholar |

Nariani TK
(1956) Leaf curl of papaya. Indian Phytopathology 9, 151–157.

Saxena S,
Hallan V,
Singh BP, Sane PV
(1998a) Leaf curl disease of Carica papaya from India may be caused by bipartite geminivirus. Plant Disease 82, 126.
| Crossref | GoogleScholarGoogle Scholar |

Saxena S,
Hallan V,
Singh BP, Sane PV
(1998b) Evidence from nucleic acid hybridization test for a geminivirus infection causing leaf curl disease of papaya in India. Indian Journal of Experimental Biology 36, 229–232.
|
CAS |

Thomas KM, Krishanaswamy CS
(1939) Leaf crinkle – A transmissible disease of Papaya. Current Science 8, 316–317.

Wang X,
Xie Y, Zhou X
(2004) Molecular characterization of two distinct Begomoviruses from papaya in China. Virus Genes 29, 303–309.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Wyatt SD, Brown JK
(1996) Detection of subgroup III geminivirus isolates in leaf extracts by degenerate primers and polymerase chain reaction. Phytopathology 86, 1288–1293.
| Crossref | GoogleScholarGoogle Scholar |

Zhang LB,
Zhou GH,
Li HP, Zhang SG
(2005) Molecular characterization of Papaya leaf curl virus infecting Carica papaya in Guangzhou and its biological test. Scientia Agricultura Sinica 38, 1805–1810.
|
CAS |