Data management pipeline for plant phenotyping in a multisite project
Kenny Billiau A , Heike Sprenger A , Christian Schudoma A , Dirk Walther A and Karin I. Köhl A BA Max Planck Institute of Molecular Plant Physiology, Am Muehlenberg 1, 14476 Potsdam OT Golm, Germany.
B Corresponding author. Email: koehl@mpimp-golm.mpg.de
Functional Plant Biology 39(11) 948-957 https://doi.org/10.1071/FP12009
Submitted: 13 January 2012 Accepted: 22 June 2012 Published: 15 August 2012
Journal Compilation © CSIRO Publishing 2012 Open Access CC BY-NC-ND
Abstract
In plant breeding, plants have to be characterised precisely, consistently and rapidly by different people at several field sites within defined time spans. For a meaningful data evaluation and statistical analysis, standardised data storage is required. Data access must be provided on a long-term basis and be independent of organisational barriers without endangering data integrity or intellectual property rights. We discuss the associated technical challenges and demonstrate adequate solutions exemplified in a data management pipeline for a project to identify markers for drought tolerance in potato. This project involves 11 groups from academia and breeding companies, 11 sites and four analytical platforms. Our data warehouse concept combines central data storage in databases and a file server and integrates existing and specialised database solutions for particular data types with new, project-specific databases. The strict use of controlled vocabularies and the application of web-access technologies proved vital to the successful data exchange between diverse institutes and data management concepts and infrastructures. By presenting our data management system and making the software available, we aim to support related phenotyping projects.
Additional keywords: controlled vocabulary, data integration, field trials, marker assisted selection, mixed schema design, ontologies.
Introduction
Modern phenotyping projects gather many data points on a large number of plants in relatively short times. Classical examples for such projects are genotype evaluation in breeding projects or marker evaluation projects (Finkel 2009; Richards et al. 2010). Typically, several groups from different institutions are involved in these studies: if field trials are involved, the project will span several years. Additionally, time constraints will arise from the restriction of the experiment to the growth period and the limitation of assessments to certain developmental stages of the plant. In projects where decisions on the subsequent experimental strategy have to be made based on data gathered by phenotyping, data have to be accessible to all collaborators rapidly and correctly for evaluation and proper decision making. Additionally, the intellectual property rights of the data owner have to be respected. Furthermore, results as well as methods used to obtain them have to be stored in a secure way to conform to rules of good scientific practice and as a prerequisite for publication.
Common obstacles for fast and reproducible data evaluation are the format of data storage, data access and documentation of the relationships between data. Primary data entry is still frequently done on paper, which remains the main and frequently only source of original data. For evaluation, data are generally entered into spreadsheet programs, which then contain original data, evaluation procedures and final results in the form of tables and graphs. The link to the method used to generate the data and the evaluation procedure often exists exclusively in the undocumented memory of the individual researchers until the results are published. Data from independent experiments are generally captured in different files. The connection between data in different files is often ambiguous, if not completely absent. Different data formats furthermore render a joint data analysis difficult, as the same parameter is given different names and expressed in different units. Joint analysis of qualitative data is often hampered by different or unclearly defined classification schemes. Data reshuffling and reformatting is time consuming and may introduce errors. Those who perform meta-analysis or modelling on the data often spend as much time in preparing the data for analysis as on the analysis itself. Thus, meta-analysis of different experiments is often conducted by comparing results of different experiments by the ‘eye-balling’ method of comparing result graphs. When the comparison is done by applying statistical methods on the basis of derived final result data like average and standard deviation instead of single values from biological replicates, statistical power is often reduced.
An additional obstacle for meta-analysis and a sensitive point in each multi-site project is data access to original data. Completely unrestricted access is obviously as bad as over-restrictive access that restricts access to original data to the owner of the data alone. The point of data access regulation must be considered at the very beginning of each multi-site project, especially when partners from industry are involved in the project.
Central data management has been state-of-the-art in highly-regulated medical and pharmacological studies for more than a decade (Nadkarni et al. 1999; Alshawi et al. 2003; Marenco et al. 2003; Bérard et al. 2012). In life sciences, the introduction of central data storage solutions has been triggered by omics studies aiming to make huge amounts of sequence information publicly available (Sherry et al. 1999; Riano-Pachon et al. 2009; Sayers et al. 2009). A positive example is also the transcript profiling by microarrays, where uploading of raw data to public databases became a standard procedure and a prerequisite for publication from the very beginning (Gollub et al. 2003; Zimmermann et al. 2004). In many cases, especially in life sciences, these solutions have been restricted to large, multinational projects and to the preservation and presentation of result data. However, even medium-sized projects may profit from central data warehouse solutions. Positive examples can be found especially in ecology and taxonomy; a reference list of respective projects has been compiled by Fabre et al. (2011). A well planned data repository improves data accessibility to all partners during the project, reduces the risk of data loss and speeds up evaluation (Alshawi Saez-Pujol et al. 2003). In the ideal case, the central data repository allows documenting all steps of the project from the methods used, the meta-information, the generated raw data and the evaluation procedures to the final result table or figure and provides long-term access to this information (Australian Plant Phenomics Facility (Li et al. 2011), Phenopsis facility (Fabre et al. 2011)).
Ecological and agricultural research projects share the problem of large variances requiring big sample size to allow statistically significant estimates. Often, a single project partner cannot produce the required number of samples within the available time. Rapidly merging data from all partners can overcome that problem. Furthermore, data have to remain accessible after the end of the project to both, the project partners and other people, to enable the experiment replication and thus publication and the practical use of the markers.
Here, we present possible solutions to the above listed challenges as exemplified in the data management concept we designed for the plant phenotyping project ‘TROST’. TROST combines field trials at 11 sites and measurements using four different analytical platforms at three different institutions. The aim of the project is to identify markers for drought tolerance in a crop plant and make these markers and the respective quantification methods available to plant breeders. The project is, therefore, a long-term project planned for ~10 years. TROST is a typical example for a phenotyping project with partners from different backgrounds (basic and applied research, industry) collaborating without hierarchies, work packages performed in different locations and the need for a comprehensive evaluation of all results during the project. Medium-sized projects of that type are funded frequently (e.g. http://foerderportal.bund.de/foekat/jsp/StartAction.do for Germany, accessed 18 June 2012) and are generally coordinated by scientists. We present our data integration solution as a model for the data management of this common project type and demonstrate how the above listed obstacles and challenges can be overcome. In addition, we make parts of the developed infrastructure available for general use.
Case study
The project named ‘TROST’ aims to identify molecular markers for drought tolerance in starch potatoes. The project involves eight research groups from four different academic institutions and seven breeding companies. Markers are to be identified from profiling metabolites and transcripts in a population of initially four check cultivars with well characterised drought tolerance. In a second phase, candidate markers are to be confirmed in a larger set of 30 potato cultivars grown under conditions ranging from fully controlled greenhouse to field conditions. Test and check cultivars are cultivated at different water supply levels in controlled environment experiments on two sites and field experiments on three sites. Additionally, all cultivars are field-cultivated at eight sites with different soil types and climate conditions in Germany. On all sites, plants are phenotyped at different times of development for performance parameters and final yield parameters and plants are sampled for metabolite and transcript analysis. Sample analysis is done by four different groups at three different institutions. The total number of samples (>7000) and the potential number of phenotyping parameters (>5000) are too high with respect to time and financial constraints to be handled exhaustively during the project. The strategy is, thus, to identify the relevant samples during the project, especially the developmental stage that delivers the most relevant samples and the most informative parameters. Therefore, raw data have to be accessible as quickly as possible during the project for modelling and decision making. Rapid evaluation of all data from all partners gathered during the first project year allows decisions concerning the optimal sampling time – reducing the time and money required for field sampling – and identification of the most relevant samples for in depth analysis of all parameters.
Data warehouse concept
The data warehouse concept utilised in the TROST project combines two already existing database systems and a newly developed database with a file server and a web server. Those data needed for modelling were put into a structured format within these databases. Data that are helpful for interpretation and replication of experiments (i.e. pictures, method descriptions), but not directly needed in calculations, are stored on the file server. The files are then linked to observations in the database. Data exchange between users is performed on web pages. The data required for statistical analysis are stored in the Plant Cultivation database system of the Max Planck Institute (MPI) Golm (Köhl et al. 2008), the Golm Metabolome Database (GMD) for metabolite profile data (Hummel et al. 2007) and the newly established Phenotyper database. All databases and servers are hosted at the MPI for Molecular Plant Physiology (Golm, Germany).
The Plant Cultivation Database system of the MPI Golm is based on a laboratory management system (LIMS). This system records information on genetic material used in the project, details of the cultivation experiments and samples (Köhl et al. 2008). The workflow uses the LIMS entities location, species, cultivar (table subspecies), plant line (table sample), plant (table aliquot), experiment (table study), project, component (table aliquot) and samples (table sample), (see Supplement 1, available as Supplementary Material to this paper). Unique IDs for each entity are provided by the Oracle sequences. Unique and interpretable entity names were designed with the syntax function of the LIMS. Within the LIMS, project data are collected in a dedicated folder that is generated by filtering for the project’s name in each entry. Thus, overview is facilitated especially for the technical staff. Each line of genetic material (i.e. seed potatoes from a certain cultivar and year) are linked to information on species, cultivar and supplier. Batches from the same cultivar are kept separate when they differ in origin, i.e. were generated by different propagation methods, in different years or by different suppliers. The plants grown in the experiments are treated as offspring of these lines. Plants are combined in experimental entities, so-called ‘cultures’, for which information on the location of cultivation, experimental procedures (fertilisation, plant protection), planting and harvest time are recorded.
Data capture
Sample information
By predefining samples in the database before they are taken, we provide a label with a unique identifier (ID). The sampling crews can link the sample ID to the sampled plants (and plant organs) by entering the plant ID in standard spreadsheet files before the harvest. The link can be established more efficiently by scanning the barcoded ID of sample label and plant label with a barcode-scanner terminal during sampling. This procedure adds a time-stamp to each component of a sample. Samples are transferred to a central storage unit with barcoded storage containers.
For each sample, a preset number of aliquots with barcoded labels are generated for analytics. Each label displays plaintext aliquot names (i.e. 134545a2) derived from the sample identifiers (i.e. 134545) to allow easy cross-reference between sample and aliquot during aliquot preparation in the laboratory. Weight and location of each aliquot is recorded in barcode-scanner terminals with the standard workflow for phenotyping (see below). Data are transferred to the LIMS, which records the processing status of each aliquot.
The meta-information about the analysed material is transferred from the LIMS system to the Phenotyper database (see below). ID data are transmitted to the meta-information table of GMD (Fig. 1) for aliquots that were metabolite-profiled. The transfer from the LIMS and the Phenotyper database is based on script-generated csv-file (Supplement 2). The meta-information comprises the identifier of the aliquot, the list of identifiers and names of the plant(s) contributing to the sample, the name of the sampled organ, the sampling date and time and the identifier of the plant’s location and culture. Thus, the necessary details for quality control and statistical analysis within the GMD are provided.
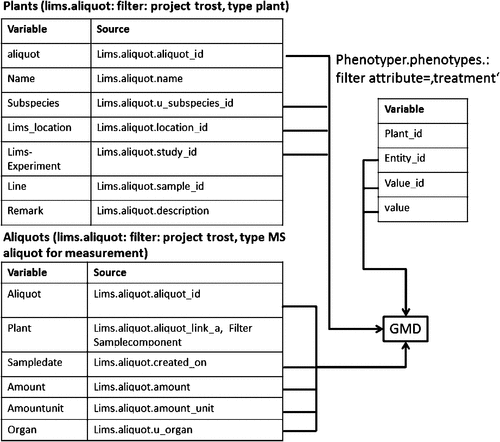
|
Likewise, all information required for evaluation, i.e. for the effects of germplasm or treatment, is directly available in the Phenotyper database, thus, the researcher does not need to be familiar with the data structure of the LIMS. Furthermore, potentially confidential data (e.g. cultivar names, test site names) are only available to those with access to the LIMS. We are, thus, using a principle that is used in medical research, where disclosure of the patient’s identity to non-medical staff is avoided by restricting some of the patient’s data to a separate database. In life sciences, breeders can keep information confidential and also share the result data with academic researchers. In this way, our system facilitates interaction between researchers from basic science and applied research and makes data available for publication as well as the practical use of the marker.
Entity-attribute-value concept in the Phenotyper database
In the Phenotyper database, entities from the LIMS, especially plants, samples and their aliquots are linked to phenotyping information in the broadest sense of the expression. In addition to measured or observed phenotypic data like plant height, FW of an aliquot, we also record features preset by the scientist (i.e. position of a plant in a row, treatment type) in the schema.
The data model of the Phenotyper is based on the entity-attribute-value concept (EAV) (Nadkarni et al. 1999) that allows linking zero to many attributes and values to each entity and object. The EAV concept is closely related to the extensible markup language concept (XML) (Dinu and Nadkarni 2007). In the EAV concept, the object, on which a test or measurement is done, i.e. a patient, a defined plant, a plot, is stored in the entity field. The attribute field contains the type of test or parameter (i.e. height), the value the quantity or quality (low, high, 150). In our model, the attribute defines the parameter that was measured and its class (for non-numeric parameters) or its unit (for numeric parameters). For numerical parameters, attribute and values are complemented by a number field. We store the information on the organ, on which the measurement has been done in the entity field. Additionally, each measurement receives a timestamp. The object, on which the measurement was performed, is identified by object type (i.e. study, sample, aliquot) and identifier. The entry in the Phenotyper table can, thus, be unambiguously linked to the information imported from the LIMS, which are stored in a conventional table. The combination of EAV concept with conventional tables of a relational database results in a mixed schema design (Dinu and Nadkarni 2007). The concept can be extended to accommodate further hierarchical levels. Thus, information on plants, cultivation experiments and sites can be stored in the same table. This contrasts to the structure of the Phenopsis database (Fabre et al. 2011), in which separate description or measurement tables are used to represent organ measurements and climate measurements in cultivation sites.
An alternative solution to represent a hierarchically nested structure has been shown in the plant trait database model (Kattge et al. 2011). Measurements taken on the same object at the same time are ‘aggregated to observations’ (Kattge et al. 2011). The relationship between the different objects is represented in a formal, mathematical model. A similar concept has been used in the Phenomics Ontology Driven Data (PODD) repository of the Australian Phenomics Facility (Li et al. 2013) to store information on plants. The PODD’s semantic web structure attaches data files to metadata objects describing their connection with controlled vocabulary.
The main advantage of the EAV-model is its open structure, which allows adding further parameters during the project without altering the database scheme. Therefore, the system is especially suitable in a project with sparse and volatile data, i.e. where the number of parameters measured during the project on an object is small compared with the number of available parameters, i.e. features that might be measured and may change during the project depending on results gained (Dinu and Nadkarni 2007). For example (Fig. 2), a surviving plant identified by a plant object identification from the plant cultivation database, can receive entries for several parameters at two scoring dates. A plant that died a few days after planting receives one entry: plant object- ‘plant id’ plant viability dead ‘date’. Changes in the number of the parameter can result from the project strategy. We will quantify expression for many thousand genes by next generation sequencing on a few samples and identify marker candidates that allow predicting tolerance. Based on the results, a few candidate genes are chosen to measure expression in many samples by qRT–PCR to obtain data for the test population in the cross-validation scheme.
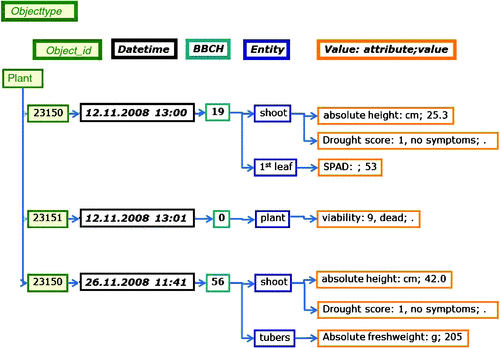
|
One of the disadvantages of the EAV model is the need to transpose the data in a columnar format, which may cause performance problems in large datasets (Marenco et al. 2003). Transformation is necessary for several statistical tests (i.e. correlation analysis), for graphical representation and to facilitate attribute-centred queries (Dinu and Nadkarni 2007). Furthermore, most scientists are used to the columnar format and so prefer this presentation format (Marenco et al. 2003).
As a result of the well defined, standardised structure, data can be efficiently evaluated with stored scripts. These scripts are written i.e. in R (R Development Core Team http://www.r-project.org, accessed 18 June 2012) and validated when the raw data are converted into results, i.e. weather data into an estimate on the drought stress to which the plants were subjected. The script is then stored on the file server together with the result file (i.e. a table or a figure) and can be used in subsequent years by all project partners to evaluate more data. The script should then be published as part of the method description in the context of the manuscript, in which the results are presented to the public. Ideally, these scripts are uploaded to one of the R script repositories (i.e. http://r-forge.r-project.org/, accessed 18 June 2012) and its link provided in the manuscript. This procedure will ‘kill two birds with one stone’, i.e. making the evaluation procedure visible to the referee and reader and making the script available to people doing similar research. Scripts uploaded without a link to their original context (i.e. a published manuscript) would, however, require additional documentation to be generally useful.
Controlled vocabulary
The efficient use of an EAV-model requires the definition of attributes and thereby the creation of an unambiguous, controlled vocabulary for all entities and attributes. For our project, entity expressions were chosen from the controlled vocabulary list derived from the Plant Ontology consortium (Jaiswal et al. 2005) and enhanced by additional terms (Supplement 3). For the class attribute ‘developmental stages’, the BBCH code for crops was used (Lancashire et al. 1991). The list of attributes was compiled from previous projects and cross checked against published ontologies on the Ontology Lookup Service (Cote et al. 2006). Additional terms were introduced for the project (Supplement 3). On the user interfaces, the terms are selectable in English and in the native language of the project partners (in our case, German). The provision of score vocabulary in the native language of technical personnel is in our experience important for method acceptance and data quality.
A well defined ontology has huge advantages for the comparison of results and their statistical evaluation (Washington et al. 2009; Mungall et al. 2010). First, they reduce ambiguous or even erroneous (e.g. typos, letter capitalisation) descriptions, which will, in turn, lead to better, more comprehensive and consistent query results. Second, ontologies represent the relationships between entities and attributes by capturing them in a logical, hierarchical and ideally, non-redundant manner. Therefore, ontologies allow categorical data (e.g. experimental conditions) to be used in actual computations such as, for example, enrichment analyses (Khatri and Dr˘aghici 2005). Thus, in medical, animal and plant sciences, ontologies are being developed and made available to the research community (Mungall 2004; Smith et al. 2004, 2005, 2007; Jaiswal et al. 2005; Yamazaki and Jaiswal 2005; Mungall et al. 2010). This includes the use of ontologies for phenotype description (Yamazaki and Jaiswal 2005; Harnsomburana et al. 2011). Despite this progress, ontologies are still underused by researchers in plant physiology or breeding. In our project, therefore, we introduced a common ontology for all partners from the very beginning. Ideally, the choice or definition of the controlled vocabulary elicits an exchange between project partners about the parameters that are to be measured, the quantification methods and the classification schemes for qualitative data. Although the compilation of the attribute lists is time-consuming and may delay the start of data gathering, the time is well spent: a uniform, well defined attribute list is one of the critical factors for a fast and correct data evaluation in multisite projects.
Data entry and retrieval
After provision of a database and definition of the controlled vocabulary, tools for the import of the data into and their retrieval from the database are to be established. For data exchange in the project, most users prefer to email spreadsheet files. Theoretically, this procedure makes the data immediately available, but practically results in a chaotic storage structure and unclear data access until the files are transferred from various email accounts to a central data repository. Additionally, data copies are left on servers along the transmission path of the emails. Thus, as a front-end, we implemented a password-protected, web-based solution that allows the user to enter the data directly or to upload files. The user management for the access regulation to the webpages is provided through the user management of the Phenotyper database. The obligatory login enables us to trace the ownerships of files and add respective tags. An overview of the data workflow is given in Fig. 3. In order to serve all parties involved, the website is available in German as well as in English.
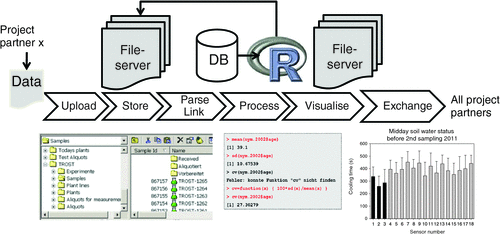
|
The web-based user interface features additional functionality enabling a user to upload several documents (Fig. 4). On the interface, the upload can be tagged with labels, to facilitate grouping and retrieving documents. Depending on the type of document, the files are either linked to entries in the database or their content is parsed into the database (Supplement 4, Fig. 1). The latter option is especially used for data in standardised formats gathered with mobile barcode scanner terminals. For data collected by the pen-and-paper method, a structured direct data entry into web templates is provided (Supplement 4, Fig. 2). Templates are especially suitable, when the number of parameters is constant throughout the project. We used this model, for example, for the entry of weather data, where a dataset consistently contains the parameters site id, date, minimum temperature, maximum temperature, precipitation and irrigation. The direct data entry on the web page facilitates quality control by predefinition of data types in the template field. It also allows validation of data according to predefined criteria (Marenco et al. 2003) directly on entry and provides a direct feedback to the user. In our experience, however, many users have concerns about direct data entries because of time-out problems in areas with low data connection width. Even more importantly, direct data entry on the webpage often means copying or even re-entering data from other sources, i.e. spreadsheet files, which is time consuming and may introduce errors (Reynolds-Haertle and McBride 1992; Gibson et al. 1994).
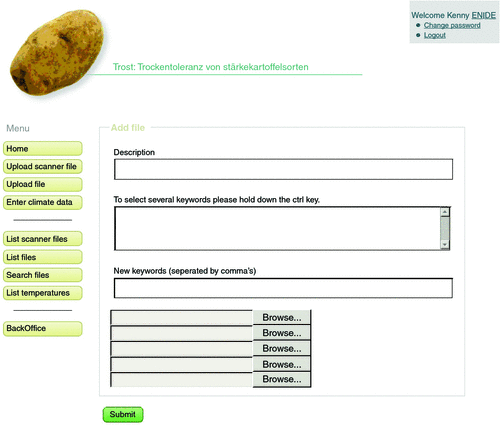
|
Most users prefer to record data manually (pen and paper) and enter them into spreadsheet files. Data transfer from spreadsheet files into the database is more complicated as spreadsheets are a priori not standardised with respect to the position of the information in the table or the data type in the table’s cells. Thus, files need to be curated manually to validate the data and the file format before their content can be uploaded to the database. This need for manual curation introduces a time delay. Efficient parsing from files into the database requires data in standard data sheets, on which parameter names match controlled vocabulary terms. To avoid a reiteration of the copying/re-entering problem mentioned above, these sheets need to be agreed on and provided in an early phase of the project. Even then, a time gap exists between gathering of data and availability in the database, which makes quality control more difficult. Much time is saved when the data is recorded in standardised formats at the time of measurement, either into a laptop or in mobile scanner terminals. Our scanner workflows allowed selecting controlled vocabulary for entities and attributes and the respective plant object onsite. The resulting files were then transferred from the scanner to the upload function of the web page. The format of these highly standardised files was recognised directly by the web page and the data parsed correctly into the Phenotyper database by a series of scripts (Supplement 5). This streamlines the process and enables us to produce consistent results as well as to flexibly adjust to any format the creator of the vocabulary provides.
Extensive validation is done before the data is entered into the database, including validation of plant and culture IDs through connection to the Oracle database of the sample and plant documentation system (LIMS). Validation includes referencing each term to the correct ontology, checking whether or not a sample exists, looking up cultures in the LIMS and making sure the measured values are stored following the set format and unit conventions.
For a smooth user experience, AJAX (Asynchronous JavaScript and XML) has been applied to automatically populate drop down menus listing previous choices. This technology avoids the reloading of a page, when the webpage interacts with the server. As this method is used by internet search tools to populate search box, most users are familiar with the tool.
In addition to files containing data that are to be stored in the database, we provided a central store for files that contain meta-information. These files will be linked to one or many database objects. For example, the pdf file with the description of the sampling procedure is linked to the samples taken according to the message. A picture showing a field plot is linked to the database object, in which the respective culture is described. Likewise, result data files, i.e. a figure or a table, are connected to the documentation of the evaluation method that was used to produce it and thus to the raw data on which the results were based. These links can be generated directly when the data are uploaded by selecting the object, to which the data are to be linked or later on. Additionally, the user can select keywords from a curated list, provide a text description to facilitate the search for a certain document and declare a document confidential. After upload, documents are stored in a protected, read-only mode on a file server to ensure long-term data integrity. Thus, the documents can no longer be modified by the user, except by changing the document status from valid to invalid. Users can search for files on the webpage and download copies of those files that have not been declared confidential. Furthermore, the user can restrict the search to valid files. The status of each document (valid/invalid) and the upload time are displayed on the result page of the document search, to facilitate identifying of the most recent version.
Technical notes
Phenotyper database
All information for the Phenotyper system is stored in a MySQL database. The graphic view of the database schema is provided in Supplement 6. A simplified version of the entity-relation diagram is represented in Fig. 5. Data import from the plant and sample database and from various file formats is done by scripts. Support libraries were written in Perl and to a limited extend in Python.
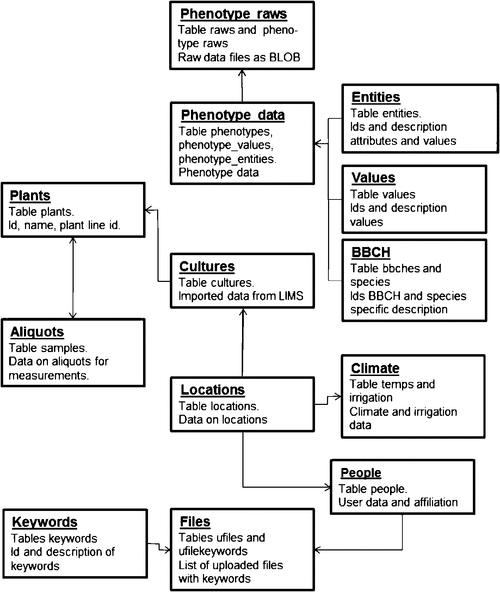
|
File server, web interface and barcode scanner
The web interface was created using the CakePHP framework, which facilitates to create scaffolds around an already existing database. The underlying technology for the front end website is PHP, JavaScript with AJAX running on an Apache server. User interfaces on the barcode scanners terminal CPT-711 L (CipherLab, Mönchengladbach, Germany) were produced with the terminal’s program generator software.
Plant and sample database
Meta-information on plants and samples taken from plants were documented in the previously described documentation system for plant resources and experiments (Köhl et al. 2008). The system is based on the commercial laboratory management system (LIMS) Nautilus (8.1, Thermo Scientific, Waltham, MA, USA) that stores data and procedures in an Oracle database (Oracle 10 g, Oracle, Redwood Shores, CA, USA). Barcode labels were designed with the function ‘report’ of the program Sybase infomaker (10.2.1, Sybase Inc., Dublin, Ireland) that links directly to the Oracle database and thus allows efficient high-throughput label printing.
Conclusion
Setting up a database solution with fileserver and webserver is admittedly considerably more work and requires more informatics knowledge than just using standard spreadsheet programs and desktop file storage. However, systems like the electronic laboratory journal Open Enventory (http://sourceforge.net/projects/enventory/, accessed 23 July 2012) indicate that scientists are increasingly prepared to acquire that knowledge and invest the time to increase efficiency. Altogether, we conclude that the time spent on setting up a data workflow for multisite project facilitates collaboration, improves data quality and speeds up evaluation within the project and ideally of other projects as well. Thus, the investment is well rewarded indeed.
Availability
A Phenotyper database (without data), its web interface and scripts can be downloaded as Supplement 5 and from the authors’ webpage (http://www-en.mpimp-golm.mpg.de/webbasedRsrc/index.html#dmp).
Acknowledgement
We thank Jürgen Gremmels and all members of the TROST consortium, especially Sylvia Seddig and Rolf Peters for helpful discussions. The work was funded by grant 22011208 of the BMELV (German Ministry for Food, Agriculture and Consumer Protection).
References
Alshawi S, Saez-Pujol I, Irani Z (2003) Data warehousing in decision support for pharmaceutical R&D supply chain. International Journal of Information Management 23, 259–268.| Data warehousing in decision support for pharmaceutical R&D supply chain.Crossref | GoogleScholarGoogle Scholar |
Bérard C, Cloutier LM, Cassivi L (2012) Evaluating clinical trial management systems: a simulation approach. Industrial Management & Data Systems 112, 146–164.
Cote R, Jones P, Apweiler R, Hermjakob H (2006) The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics 7, 97
Dinu V, Nadkarni P (2007) Guidelines for the effective use of entity-attribute-value modeling for biomedical databases. International Journal of Medical Informatics 76, 769–779.
| Guidelines for the effective use of entity-attribute-value modeling for biomedical databases.Crossref | GoogleScholarGoogle Scholar |
Fabre J, Dauzat M, Negre V, Wuyts N, Tireau A, Gennari E, Neveu P, Tisne S, Massonnet C, Hummel I, Granier C (2011) PHENOPSIS DB: an information system for Arabidopsis thaliana phenotypic data in an environmental context. BMC Plant Biology 11, 77
| PHENOPSIS DB: an information system for Arabidopsis thaliana phenotypic data in an environmental context.Crossref | GoogleScholarGoogle Scholar |
Finkel E (2009) With ‘Phenomics,’ plant scientists hope to shift breeding into overdrive. Science 325, 380–381.
| With ‘Phenomics,’ plant scientists hope to shift breeding into overdrive.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXpt1GjtL4%3D&md5=e770a9a60203a82cc9b4d4ed4e500658CAS |
Gibson D, Harvey AJ, Everett V, Parmar MKB (1994) Is double data-entry necessary – the CHART trials. Controlled Clinical Trials 15, 482–488.
| Is double data-entry necessary – the CHART trials.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2M7lsVSjsA%3D%3D&md5=0007325534b43a860693cc51428bd092CAS |
Gollub J, Ball CA, Binkley G, Demeter J, Finkelstein DB, Hebert JM, Hernandez-Boussard T, Jin H, Kaloper M, Matese JC, Schroeder M, Brown PO, Botstein D, Sherlock G (2003) The Stanford microarray database: data access and quality assessment tools. Nucleic Acids Research 31, 94–96.
| The Stanford microarray database: data access and quality assessment tools.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXhvFSnsrY%3D&md5=3b91922c324661cbbfa3e196667fe3c7CAS |
Harnsomburana J, Green JM, Barb AS, Schaeffer M, Vincent L, Shyu CR (2011) Computable visually observed phenotype ontological framework for plants. BMC Bioinformatics 12, 260
| Computable visually observed phenotype ontological framework for plants.Crossref | GoogleScholarGoogle Scholar |
Hummel J, Selbig J, Walther D, Kopka J (2007) The Golm Metabolome Database: a database for GC-MS based metabolite profiling. Topics in Current Genetics 18, 75–95.
| The Golm Metabolome Database: a database for GC-MS based metabolite profiling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXntlWqsg%3D%3D&md5=1f725e0e3d52e17f85a8c19e5a49ecbdCAS |
Jaiswal P, Avraham S, Ilic K, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M, Stein L, Stevens P, Vincent L, Ware D, Zapata F (2005) Plant ontology (PO): a controlled vocabulary of plant structures and growth stages. Comparative and Functional Genomics 6, 388–397.
| Plant ontology (PO): a controlled vocabulary of plant structures and growth stages.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xit1Knur0%3D&md5=c2c6f0819dca1bec4acf49262dd4e38eCAS |
Kattge J, Ogle K, Bönisch G, Díaz S, Lavorel S, Madin J, Nadrowski K, Nöllert S, Sartor K, Wirth C (2011) A generic structure for plant trait databases. Methods in Ecology and Evolution 2, 202–213.
| A generic structure for plant trait databases.Crossref | GoogleScholarGoogle Scholar |
Khatri P, Dr˘aghici S (2005) Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics 21, 3587–3595.
| Ontological analysis of gene expression data: current tools, limitations, and open problems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXpvFGqsb0%3D&md5=1129574b2b87793e034b9f2c9cef377dCAS |
Köhl KI, Basler G, Luedemann A, Selbig J, Walther D (2008) A plant resource and experiment management system based on the Golm Plant Database as a basic tool for omics research. Plant Methods 4, 11
| A plant resource and experiment management system based on the Golm Plant Database as a basic tool for omics research.Crossref | GoogleScholarGoogle Scholar |
Lancashire PD, Bleiholder H, Van Den Boom T, Landgeluddeke P, Strauss R, Weber E, Witzenberger A (1991) A uniform decimal code for growth stages of crops and weeds. Annals of Applied Biology 119, 561–601.
| A uniform decimal code for growth stages of crops and weeds.Crossref | GoogleScholarGoogle Scholar |
Li Y-F, Kennedy G, Ngoran F, Wu P, Hunter J (2011) An ontology-centric architecture for extensible scientific data management systems. Future Generation Computer Systems in press.
Marenco L, Tosches N, Crasto C, Shepherd G, Miller P, Nadkarni P (2003) Achieving evolvable web-database bioscience applications using the EAV/CR framework: recent advances. Journal of the American Medical Informatics Association 10, 444–453.
| Achieving evolvable web-database bioscience applications using the EAV/CR framework: recent advances.Crossref | GoogleScholarGoogle Scholar |
Mungall CJ (2004) Obol: integrating language and meaning in bio-ontologies. Comparative and Functional Genomics 5, 509–520.
| Obol: integrating language and meaning in bio-ontologies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtFertbo%3D&md5=034da8f487835fda6a458dd2b5582f5aCAS |
Mungall C, Gkoutos G, Smith C, Haendel M, Lewis S, Ashburner M (2010) Integrating phenotype ontologies across multiple species. Genome Biology 11, R2
| Integrating phenotype ontologies across multiple species.Crossref | GoogleScholarGoogle Scholar |
Nadkarni PM, Marenco L, Chen R, Skoufos E, Shepherd G, Miller P (1999) Organization of heterogeneous scientific data using the EAV/CR representation. Journal of the American Medical Informatics Association 6, 478–493.
| Organization of heterogeneous scientific data using the EAV/CR representation.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3c%2FksVajtQ%3D%3D&md5=cda5c35da840cc42eeb062ca17ccbda0CAS |
Reynolds-Haertle RA, McBride R (1992) Single vs double data entry in CAST. Controlled Clinical Trials 13, 487–494.
| Single vs double data entry in CAST.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK3s7gsFKhsg%3D%3D&md5=746d78097dadf941954cf0221699d1fcCAS |
Riano-Pachon DM, Nagel A, Neigenfind J, Wagner R, Basekow R, Weber E, Mueller-Roeber B, Diehl S, Kersten B (2009) GabiPD: the GABI primary database – a plant integrative ‘omics’ database. Nucleic Acids Research 37, D954–D959.
| GabiPD: the GABI primary database – a plant integrative ‘omics’ database.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsFejtL%2FO&md5=81e7f28134b9e3761173c20aa4cf9a42CAS |
Richards RA, Rebetzke GJ, Watt M, Condon AG, Spielmeyer W, Dolferus R (2010) Breeding for improved water productivity in temperate cereals: phenotyping, quantitative trait loci, markers and the selection environment. Functional Plant Biology 37, 85–97.
| Breeding for improved water productivity in temperate cereals: phenotyping, quantitative trait loci, markers and the selection environment.Crossref | GoogleScholarGoogle Scholar |
Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E, Ye J (2009) Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 37, D5–D15.
| Database resources of the National Center for Biotechnology Information.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsFejt7fN&md5=446409177ebdafeeb885011b69a01a5bCAS |
Sherry ST, Ward MH, Sirotkin K (1999) dbSNP – Database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Research 9, 677–679.
Smith CL, Goldsmith CA, Eppig JT (2004) The mammalian phenotype ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biology 6, R7
| The mammalian phenotype ontology as a tool for annotating, analyzing and comparing phenotypic information.Crossref | GoogleScholarGoogle Scholar |
Smith B, Ceusters W, Kohler J, Kumar A, Lomax J, Mungall CJ, Neuhaus F, Rector A, Rosse C (2005) Relations in biomedical ontologies. Genome Biology 6, R46
| Relations in biomedical ontologies.Crossref | GoogleScholarGoogle Scholar |
Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S-A, Scheuermann RH, Shah N, Whetzel PL, Lewis S (2007) The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature Biotechnology 25, 1251–1255.
| The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXht1Oru7zK&md5=2ab5bcc4ac71fade4a3425ad1d976e8cCAS |
Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE (2009) Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biology 7, e1000247
| Linking human diseases to animal models using ontology-based phenotype annotation.Crossref | GoogleScholarGoogle Scholar |
Yamazaki Y, Jaiswal P (2005) Biological ontologies in rice databases. An introduction to the activities in gramene and oryzabase. Plant & Cell Physiology 46, 63–68.
| Biological ontologies in rice databases. An introduction to the activities in gramene and oryzabase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXht1eqt7w%3D&md5=252aa64009b566b3aad883ad5a45f1d3CAS |
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632.
| GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXnvFOru74%3D&md5=756aecd9238590fd8f3a3c36afca5916CAS |


