Seed storage albumins: biosynthesis, trafficking and structures
Joshua S. Mylne A D , Ikuko Hara-Nishimura B and K. Johan Rosengren CA The University of Western Australia, School of Chemistry and Biochemistry and ARC Centre of Excellence in Plant Energy Biology, 35 Stirling Highway, Crawley, Perth, WA 6009, Australia.
B Department of Botany, Graduate School of Science, Kyoto University, Kitashirakawa-oiwake cho Sakyo-ku, Kyoto, 606-8502, Japan.
C The University of Queensland, School of Biomedical Sciences, Brisbane, Qld 4072, Australia.
D Corresponding author. Email: joshua.mylne@uwa.edu.au
This review originates from the Peter Goldacre Award 2012 of the Australian Society of Plant Scientists that was received by the first author.
Functional Plant Biology 41(7) 671-677 https://doi.org/10.1071/FP14035
Submitted: 24 January 2014 Accepted: 24 March 2014 Published: 6 May 2014
Abstract
Seed storage albumins are water-soluble and highly abundant proteins that are broken-down during seed germination to provide nitrogen and sulfur for the developing seedling. During seed maturation these proteins are subject to post-translational modifications and trafficking before they are deposited in great quantity and with great stability in dedicated vacuoles. This review will cover the subcellular movement, biochemical processing and mature structures of seed storage napins.
Additional keywords: asparaginyl endo-peptidase, napin, seed storage protein, vacuolar processing enzyme, 2S albumin.
Napin-type seed storage albumin
Albumin is a generic name for a biochemical property and was originally attributed to any protein that was soluble in water. In plant seeds, there is a proportion of proteins that are soluble in distilled water and these seed storage albumins have traditionally been characterised and referred to by their sedimentation co-efficient (i.e. 2S albumins) and their similarity to napin – the first well studied seed storage albumin (i.e. napins). The seed storage albumins of model plant Arabidopsis thaliana (L. Heynh.) have also been referred to as arabins (Heath et al. 1986).
The remainder of the seed storage proteins require addition of salt to dissolve and these are termed globulins (also known as legumins, cruciferins). Many of the processing events and trafficking pathways described here for albumins are similar or identical for these seed globulins.
Seed storage albumins were reviewed comprehensively by Peter Shewry and colleagues in the mid-late 1990s (Shewry et al. 1995; Shewry and Pandya 1999) and more recently in the context of what makes 2S albumin allergenic (Moreno and Clemente 2008). The present review focusses on the napin-type seed storage albumins and encompasses their biosynthesis, trafficking, proteolytic maturation as well as their mature tertiary structures.
The preproalbumin
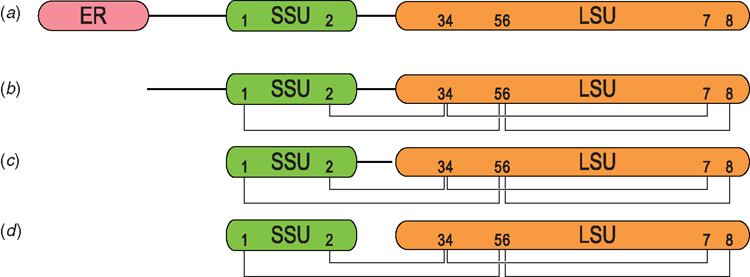
Plants usually contain several to many preproalbumin genes that are often intronless and found in tandem or arrays. Preproalbumins typically constitute an endoplasmic reticulum (ER) signal domain and a pro-domain, followed by the albumin domain. After the preproalbumin has its ER signal removed, it is referred to as proalbumin. The proalbumin is subsequently subjected to proteolytic processing leading to the mature albumin (Fig. 1). Here we follow the steps involved in the maturation of preproalbumin into albumin and the concurrent changes in subcellular location as this trafficking and post-translational ballet ensues.
Processing from preproalbumin to folded proalbumin
During translation the ER signal peptide protrudes from the ribosome large subunit and is recognised by the Signal Recognition Particle. The binding of the Signal Recognition Particle slows protein synthesis and consequently folding so that the ribosome may be taken to the ER membrane. Once there, protein synthesis resumes and the signal peptide is removed co-translationally by signal peptidases (reviewed by Paetzel et al. 2002). By combining in vitro translation and pulse-chase experiments with seed cotyledons, the precise cleavage point for ER signal sequence removal can be determined, as demonstrated for a pumpkin 2S albumin (Hara-Nishimura et al. 1993).
The disulfide bonds of proalbumins are the most obviously conserved feature and it is in the ER lumen, with its high oxidative redox potential that disulfide bonds form. The correct folding and formation of the native disulfide bonds is a critical step in the production of any disulfide rich protein, and often requires assistance from chaperones or disulfide isomerases. Plants express a wide range of protein disulfide isomerases and some have been identified to play a key role in the folding of seed albumins (Onda et al. 2011). From the ER, proalbumins are destined for protein storage vacuoles. The protein storage vacuole differs substantially from the better known lytic vacuole of vegetative cells, which occupy the majority of the plant cell, have an acidic pH, store ions and secondary metabolites as well as play active roles in water balance or turnover of proteins, lipids and carbohydrates. The protein storage vacuoles, to which albumins are transported, have a more neutral pH than lytic vacuoles, are smaller in size and as their name implies – are rich in storage proteins. The pH of protein storage vacuoles was shown to be ~6 during the late torpedo embryo stage and a pH of ~4.9 in the mature embryo. During storage protein deposition and co-incident embryo development the luminal pH of the protein storage vacuole is thought to gradually drop from ~5.5 to ~4.9 (Otegui et al. 2006).
A host of proteins that are essential for the proper deposition of seed-storage proteins into storage vacuoles have been identified. Some of the first mutants affecting sorting were identified using reverse genetic approaches targeting proteins found to be abundant in vesicles rich in storage protein precursors. The abundant pumpkin protein PV72 was discovered for its binding of peptide fragments of proalbumin (Shimada et al. 1997); this allowed the subsequent identification of seven homologues in Arabidopsis and genetic defects in one of these homologues, VACUOLAR SORTING RECEPTOR 1 was shown to cause incorrect sorting of storage proteins (Shimada et al. 2003a). Another productive route to finding proteins involved in sorting was Arabidopsis mutant screens that looked for an accumulation of incompletely processed storage proteins. This approach led to identification of the maigo (mag) mutants (Li et al. 2006; Shimada et al. 2006; Takahashi et al. 2010; Takagi et al. 2013). The MAIGO1 protein was shown to be a homologue of a component of the retromer complex and this allowed the identification of other seed storage impaired mutants by reverse genetic approaches with Arabidopsis homologues of the retromer complex such as VPS35 (Yamazaki et al. 2008). More recently, TAP-tagged MAIGO2 was used to pull down three interacting proteins (MIP1-MIP3) of differing identities, which were all confirmed through individual mip knockout phenotypes to be essential for storage protein transport. This work implies that MAIGO2 and at least these three MIP proteins constitute a protein complex that transports seed storage proteins (Li et al. 2013). MAIGO4 functions as a Golgi-tethering factor (Takahashi et al. 2010), whereas the recently published MAIGO5 is a putative orthologue of yeast Sec16 and shown to have a functional role in protein export from the ER (Takagi et al. 2013).
At present, it is thought there are two routes for storage protein transport from the ER to the protein storage vacuole; one is receptor-mediated and via the Golgi apparatus whereas the other bypasses the Golgi apparatus and is termed aggregation sorting. The latter route involves ER-derived vesicles rich in storage protein precursors that were originally found in developing pumpkin and designated as precursor-accumulating (PAC) vesicles (Hara-Nishimura et al. 1993).
To monitor the trafficking of seed storage proteins as well as their processing machinery, Otegui et al. (2006) raised antibodies for ‘spacer’ regions of Arabidopsis SEED STORAGE ALBUMIN 2 (SESA2, At4g27150, At2S-2). Two peptide antigens were used: one for a region between the SESA2 ER signal and the small subunit and the other for the region joining the small and large SESA2 subunits. Immunolabelling with these antibodies would thus track preproalbumin and proalbumin. In addition, antibodies for the seed storage processing proteases ASPARAGINYL ENDOPEPTIDASE 2 (AEP2, At1g62710, β-VPE), the VACUOLAR SORTING RECEPTOR 1, the aspartic protease A1, mature albumin and globulin as well as others were employed. This approach showed that although globulins and albumins were found together, the processing machinery that is also ER-targeted was found in regions distinct from those containing the storage proteins themselves. The immuno-labelling as well as subcellular fractionation experiments both indicated that storage proteins and their proteases are sorted into different types of vesicle. These two vesicles later fuse and become what are termed multi-vesicular bodies, thereby allowing the storage proteins and their proteolytic processing machinery to meet each other before merging with the protein storage vacuole (Otegui et al. 2006).
Maturation of proalbumin to albumin
Proalbumins and the proteases that process them are both ER-targeted, but thought to occupy separate regions of the Golgi cisternae, resulting in them being packaged separately into vesicles and not coming into contact with each other until they are delivered to multi-vesicular bodies (Otegui et al. 2006). Within the multi-vesicular bodies the proalbumin is matured by proteases, the most important being asparaginyl endopeptidase. AEP typically cuts proalbumin at two positions: the proto-N-termini of its small and large albumin subunits. These cleavages change the internally disulfide-bonded proalbumin monomer into a heterodimer held together by usually four disulfide bonds. Not all albumins are cleaved into small and large subunits; e.g. the sunflower SFA8 albumin has been found to be a monomer in its mature form (Kortt et al. 1991). Subsequent to the action of AEP, the activity of an aspartic exo-protease has been proposed to further process albumin by trimming the C-terminal regions of the cleaved propetides (Hiraiwa et al. 1997). Two regions may be trimmed from proalbumin; the C-terminal tail of the large albumin subunit and the C-terminal tail of the small subunit.
The N-termini of the matured small and large albumin subunits often possess residues or post-translational modifications that are thought to protect them from degradation by aminopeptidases. The most common N-terminal residue for mature albumin subunits is proline, which effectively ‘caps’ the mature proteins. N-terminal prolines have been shown for the monomeric SFA8 (Kortt et al. 1991) and both the small and large subunits of PawS1 and PawS2 (Mylne et al. 2011). Another common N-terminal residue is pyro-Glu, which is caused by the side chain cyclisation of either glutamic acid or glutamine. The conversion of Gln to pyro-Glu is a deamination reaction that confers resistance to degradation by aminopeptidases (Schilling et al. 2008). Pyrolated N-termini have been shown for subunits of mabinlin II (Liu et al. 1993) and an albumin from castor bean (Sharief and Li 1982).
Asparaginyl endo-peptidase
AEP (also known as vacuolar processing enzyme, VPE or legumain) was first identified from castor bean seeds in the early 1990s for its ability to cleave after Asn (Hara-Nishimura et al. 1991). Retrospectively, it was clear that AEP was responsible for the Cys protease activity that had been observed earlier in castor bean (Harley and Lord 1985) and pumpkin seeds (Hara-Nishimura and Nishimura 1987). Several subsequent studies showed that AEP was very capable of maturing seed storage proteins in vitro and had a strong preference for Asn and, to a lesser extent, for Asp. In proteins matured by AEP, the most commonly observed target was cleavage between Asn-Pro bonds. The importance of AEP for seed storage protein processing was most strikingly shown in Arabidopsis where loss-of-function genetic mutants were used to unpick AEP function. Extensive screening for mutations affecting seed storage protein profiles not only revealed many genes involved in in seed storage trafficking, but also found lesions in Arabidopsis AEP2 (At1g62710, β-VPE) (Shimada et al. 2003b). Arabidopsis aep2 mutants had mild defects in seed storage protein profile (Gruis et al. 2002; Shimada et al. 2003b). Single mutation in the other three AEP genes did not give noticeable changes in seed protein profile, but when lesions in AEP genes were pyramided the resulting plants had dramatically perturbed seed storage protein profiles (Shimada et al. 2003b; Gruis et al. 2004). Quadruple aep null plants had a dramatically different seed protein profile, but this defective processing did not affect seed germination or the adult plant phenotype, which makes one wonder what the purpose of all this post-translational processing is? A close analysis of the proteins in aep null seeds showed that in the absence of AEP, rather than remain completely unprocessed the proalbumins (and proglobulins) were processed improperly and inaccurately by other proteases that otherwise played little or no role in albumin maturation (Shimada et al. 2003b; Gruis et al. 2004). Proteomic analysis of the albumin in Arabidopsis wild type and aep null plants also showed that one of the four Arabidopsis SEED STORAGE ALBUMIN (SESA) proteins was not fully dependent upon AEP for its maturation. Specifically the large albumin subunit of SESA3 (At4g27160, At2S-3) is N-terminally matured in an AEP-independent fashion although by which protease is not clear (Shimada et al. 2003b; Gruis et al. 2004).
Although its importance in seed storage protein processing under standard growth conditions is the focus here, AEP has an important biological function in caspase-like inducible cell death as part of a resistance response to the detection of viral entry (Hatsugai et al. 2004) or fungal toxins (Kuroyanagi et al. 2005). Biochemically, AEP has also been found to readily perform a transpeptidation or protein ligation reaction in vitro (Min and Jones 1994) and more recently, AEP has been demonstrated to be involved in the biosynthesis of several different types of cyclic plant peptides (Mylne et al. 2012).
Albumin structure
The first primary structure for a processed and mature napin was presented by Ericson et al. (1986). In this study the authors both cloned a transcript from rapeseed for a napin-type albumin precursor and purified the corresponding mature napin. They reduced and alkylated the napin to separate the small and large subunits and partly sequenced them to describe the multiple processing events and region removed from the precursor to result in a mature, heterodimeric albumin.
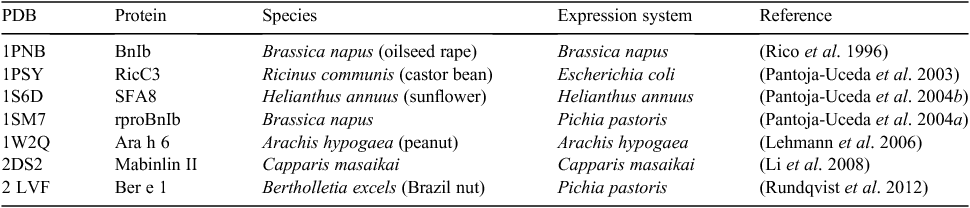
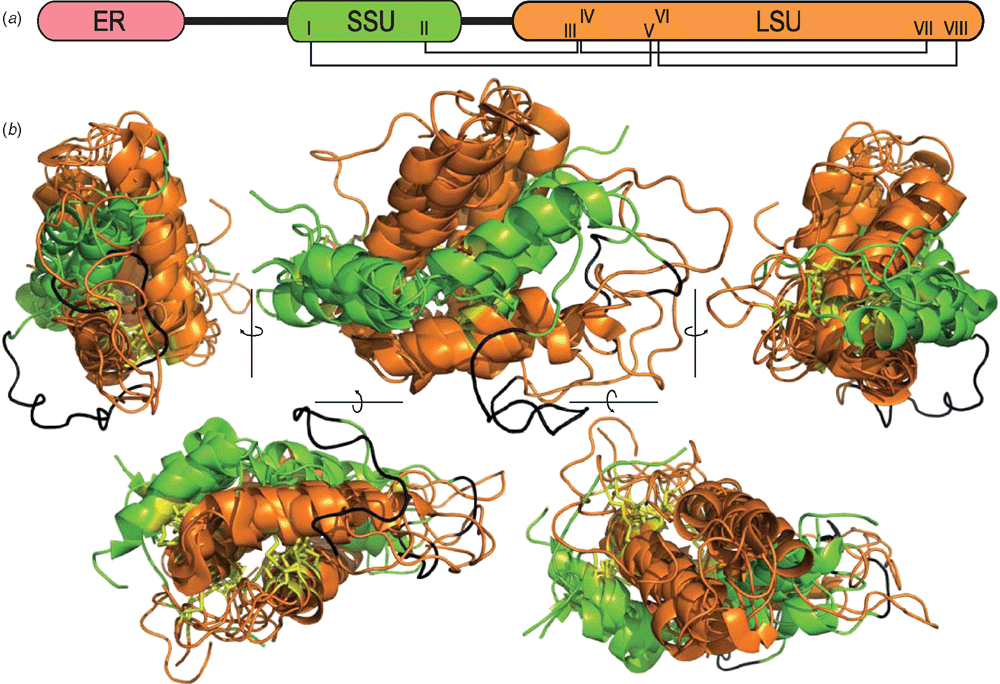
The tertiary structure was solved for the prototypic napin by Rico et al. (1996). Since then data for six more napin-type seed storage albumin have been deposited in the Protein Data Bank (PDB). In all, four of the structures are native proteins extracted from seeds, whereas the remaining three were expressed recombinantly in Escherichia coli or Pichia pastoris (Table 1; Fig. 2). The structure of mabinlin II was resolved by X-ray crystallographic analysis whereas the remainder of the structures were solved using solution NMR spectroscopy techniques. The latter technique is particularly useful for studying protein dynamics, and Rundqvist et al. (2012) used their expression system to produce isotopically labelled Ber e 1, allowing a detailed description of its internal motion.
|
|
Albumins typically possess eight Cys residues that show the connectivity: I-V, II-III, IV-VII, VI-VIII. The number of residues between CysI and CysII in the small albumin subunit ranges between 9–14 residues whereas the CysIII and CysIV at the beginning of the large subunit are a highly conserved di-Cys pair. CysV and CysVI are also highly conserved and flank a single residue whereas CysVII and CysVIII vary in their position and may be separated by 3–7 residues. Although connectivity of the four disulfide bonds is conserved among all albumins, Arachis hypogaea Ara h 6 (1W2Q) has an extra disulfide bond between Cys86 and Cys126 and there are many other examples of gene-predicted albumin precursors with 10 Cys residues that could similarly form five disulfide bonds.
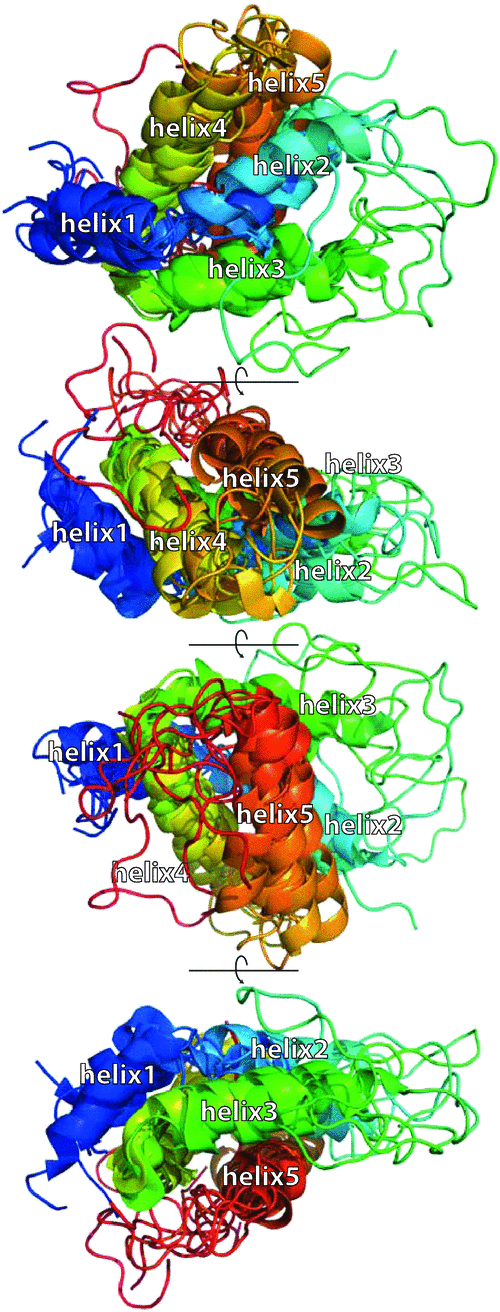
The structures all comprise five helices that are closely packed into a bundle creating a similar global fold (Fig. 3). The small albumin subunit possesses two helices that adopt a ‘V’ shape by a bend in the loop between helix1 and helix2. Within the large albumin subunit, helix3 and helix4 adopt an open hairpin conformation and this open hairpin is saddled by the V-shaped small albumin subunit. Helix5 loops back along helix4 in a hairpin and although some contact is made between helix5 and the end of helix2 of the small subunit, most of its contacts are with helix3 and helix4 of the large albumin subunit. The combination of the disulfide bonds and the compact fold makes the albumins extremely stable and resistant towards proteolytic degradation and heat (Rundqvist et al. 2012).
|
A cleavage between large and small subunit was found not to cause a significant change in structure for BnIb. The native BnIb structure (Rico et al. 1996) was compared with a recombinant form called rproBnIb expressed in Pichia pastoris and identical in primary structure except for Ser-Glu-Asn, which joins the small to large subunits (Pantoja-Uceda et al. 2004a).
Evolution of albumins
Provided they maintain their conserved Cys pattern, albumins appear to be free to evolve. Within a single plant species there can be considerable variation in sequence among albumins. The common sunflower possesses six known albumin genes HaG5 (Allen et al. 1987), HaB1B2 (GenBank AJ275962), pHAO (Thoyts et al. 1996), SFA8 (Kortt et al. 1991), PawS1 and PawS2 (Mylne et al. 2011). HaG5 and HaB1B2 have a general structure shown Fig. 1, but they each encode two mature albumins. SFA8 encodes a mature albumin that is monomeric and the PawS1 and PawS2 genes each encode a typical heterodimeric albumin as well as a small cyclic peptide.
The PawS1 and PawS2 genes from sunflower provide interesting examples of innovations from within preproalbumin. These two genes encode otherwise normal napin-type albumins, but within the pro-region between the ER signal and the small albumin subunit is a region that, through the processing by AEP forms small cyclic peptides that have a single disulfide bond (Mylne et al. 2011). The 14-residue peptide buried in PawS1 was isolated previously and is called Sunflower Trypsin Inhibitor 1 (Luckett et al. 1999) and as its name suggests it is a potent (Ki 0.1 nM) inhibitor of trypsin. PawS2 was found by homology to PawS1 and encoded by its sequence is a 12-residue peptide of similar sequence to SFTI-1 that is not trypsin-inhibitory. This SFTI-Like 1 (SFT-L1) peptide is in a similar region to SFTI-1, but is preceded directly by the ER signal sequence (Mylne et al. 2011). There are interesting parallels between this hijacked preproalbumin and PV100; a precursor for a larger storage protein called vicilin in pumpkin seeds. The action of AEP was responsible for cleaving PV100 into vicilin as well as three Arg-rich peptides and two Cys-rich peptides, one of which inhibits trypsin (Yamada et al. 1999).
Questions to be answered
Albumins are rapidly evolving proteins that are produced so they may be broken down during germination. The albumins for which structures have been solved have a structurally similar, five-helix fold. Apart from the conserved Cys residues and a general richness in Gln, albumin sequences are not conserved, but fold into a common structure implying the five-helix structure is advantageous for such highly abundant proteins to fold well and remain stable during long-term storage. It seems strange that the perturbations in processing caused by the loss of the major processing enzyme AEP have no effect on seed germination. Although all storage proteins in AEP knockouts are mis-processed and the gross seed protein profile is greatly altered, the disulfide bonds in each protein are likely to form correctly in these mis-processed proteins as disulfide bond formation and albumin folding precedes the maturation by AEP. The structures of BnIb (1PNB) and rproBnIb (1SM7) are identical despite rproBnIb possessing a sequence that is usually processed out to produce mature small and large BnIb subunits, so a lack of processing by AEP may have little effect on the overall structure.
Storage proteins may also be an untapped source of new proteins. Most plants have several or many genes for their seed storage proteins that, combined with the observation that they can vary their sequences greatly, make storage proteins potentially interesting to explore for novel peptides and bioactivities. The sunflower PawS proteins and PV100 are good examples, but there could be many more waiting to be found. Most expressed sequence tag or next-generation sequencing projects study green tissues and avoid seeds because of their less diverse transcriptome. The current table of over 1300 plant samples sequences held by The 1000 Plants Initiative (www.onekp.com, accessed 7 April 2014) has only three samples that list seeds as their tissue type. With the lowering costs for next-generation sequencing and development of better tools to analyse the data, sequencing seed transcriptomes is more viable than ever.
Finally, studying the evolution of seed storage proteins is daunting because of their rapid evolution. What makes a ‘good’ albumin? Is it just long-term storage or is it a store that is ideally suited for the subsequent mobilisation as the seed germinates? Seed storage proteins are arguably one of the most important sources of protein for humans and their abundance made them the subject of many of the first studies in plant protein biochemistry. Perhaps it is time to redouble our efforts using the panoply of molecular and biochemical tools and techniques now available?
Acknowledgements
JSM was supported by an Australian Research Council (ARC) Future Fellowship (FT120100013). Work on this topic is supported by ARC grant DP120103369 to JSM and KJR. JSM is indebted to David Craik (University of Queensland) for the introduction and mentoring in the field of protein biochemistry. JSM would also like to thank Ian Small (University of Western Australia) for ongoing support. IH-N acknowledges support by a Specially Promoted Research of Grant-in-Aid from the Japan Society for the Promotion of Science (22000014). KJR is an ARC Future Fellow (FT130100890).
References
Alcocer MJC, Murtagh GJ, Bailey K, Dumoulin M, Sarabia Meseguer A, Parker MJ, Archer DB (2002) The disulphide mapping, folding and characterisation of recombinant Ber e 1, an allergenic protein, and SFA8, two sulphur-rich 2 S plant albumins. Journal of Molecular Biology 324, 165–175.| The disulphide mapping, folding and characterisation of recombinant Ber e 1, an allergenic protein, and SFA8, two sulphur-rich 2 S plant albumins.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XotlWmsb8%3D&md5=cb669d8c1250f0f8b398325e1fcf5531CAS |
Allen R, Cohen E, Vonder Haar R, Adams C, Ma D, Nessler C, Thomas T (1987) Sequence and expression of a gene encoding an albumin storage protein in sunflower. Molecular & General Genetics 210, 211–218.
| Sequence and expression of a gene encoding an albumin storage protein in sunflower.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1cXhvVWiuro%3D&md5=b60102ad11e4f53bc96ae034056d8e00CAS |
Ericson ML, Rödin J, Lenman M, Glimelius K, Josefsson LG, Rask L (1986) Structure of the rapeseed 1.7 S storage protein, napin, and its precursor. Journal of Biological Chemistry 261, 14 576–14 581.
Gruis DF, Selinger DA, Curran JM, Jung R (2002) Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases. The Plant Cell 14, 2863–2882.
| Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XovF2qurc%3D&md5=ff8bedd63438ac0f0cd20951a27df19eCAS | 12417707PubMed |
Gruis D, Schulze J, Jung R (2004) Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. The Plant Cell 16, 270–290.
| Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXosVektg%3D%3D&md5=b03139f3dec21cd2b1aca261930d3a59CAS | 14688293PubMed |
Hara-Nishimura I, Nishimura M (1987) Proglobulin processing enzyme in vacuoles isolated from developing pumpkin cotyledons. Plant Physiology 85, 440–445.
| Proglobulin processing enzyme in vacuoles isolated from developing pumpkin cotyledons.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1cXptVCqsQ%3D%3D&md5=5c99410a9e21d58f73cef8a113141a90CAS | 16665717PubMed |
Hara-Nishimura I, Inoue K, Nishimura M (1991) A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Letters 294, 89–93.
| A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38XhtVWmsLo%3D&md5=603a0192b6b6dd38a3fb6eae279ebd11CAS | 1743299PubMed |
Hara-Nishimura I, Takeuchi Y, Inoue K, Nishimura M (1993) Vesicle transport and processing of the precursor to 2S albumin in pumpkin. The Plant Journal 4, 793–800.
| Vesicle transport and processing of the precursor to 2S albumin in pumpkin.Crossref | GoogleScholarGoogle Scholar |
Harley SM, Lord MJ (1985) In vitro endoproteolytic cleavage of castor bean lectin presursors. Plant Science 41, 111–116.
| In vitro endoproteolytic cleavage of castor bean lectin presursors.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL28XhtVKiu70%3D&md5=7ef5ad3856a31201ceb4d75671a4e688CAS |
Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305, 855–858.
| A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmt1Krsbw%3D&md5=22be6d427ca0d5a6939afe30c417c70aCAS | 15297671PubMed |
Heath J, Weldon R, Monnot C, Meinke D (1986) Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta 169, 304–312.
| Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2sXkslaltw%3D%3D&md5=6cb2a7bba436a3f175521d91a2d2e742CAS | 24232640PubMed |
Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I (1997) An aspartic endopeptidase is involved in the breakdown of propeptides of storage proteins in protein-storage vacuoles of plants. European Journal of Biochemistry 246, 133–141.
| An aspartic endopeptidase is involved in the breakdown of propeptides of storage proteins in protein-storage vacuoles of plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXjslSjtb8%3D&md5=c3fa8219a06397c4beddf5331aa93fa5CAS | 9210475PubMed |
Irwin S, Keen J, Findlay JC, Lord JM (1990) The Ricinus communis 2S albumin precursor: a single preproprotein may be processed into two different heterodimeric storage proteins. Molecular & General Genetics 222, 400–408.
| The Ricinus communis 2S albumin precursor: a single preproprotein may be processed into two different heterodimeric storage proteins.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3cXls1Klsr0%3D&md5=5554d486f8b7e4472eb894f51bbb5bd0CAS |
Kortt AA, Caldwell JB, Lilley GG, Higgins TJV (1991) Amino acid and cDNA sequences of a methionine-rich 2S protein from sunflower seed (Helianthus annuus L.). European Journal of Biochemistry 195, 329–334.
| Amino acid and cDNA sequences of a methionine-rich 2S protein from sunflower seed (Helianthus annuus L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXks1Kqt7o%3D&md5=85e2aaf0231f869a27242589bcd452a9CAS | 1997318PubMed |
Kuroyanagi M, Yamada K, Hatsugai N, Kondo M, Nishimura M, Hara-Nishimura I (2005) Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. Journal of Biological Chemistry 280, 32 914–32 920.
| Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVWiurrN&md5=4a0bb841a2345a4a8d1d7c33cdd6040aCAS |
Lehmann K, Schweimer K, Reese G, Randow S, Suhr M, Becker W-M, Vieths S, Rösch P (2006) Structure and stability of 2S albumin-type peanut allergens: implications for the severity of peanut allergic reactions. Biochemical Journal 395, 463–472.
| Structure and stability of 2S albumin-type peanut allergens: implications for the severity of peanut allergic reactions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjtlCgsLs%3D&md5=f7fff07e575780316e6b14bfa3ad74abCAS | 16372900PubMed |
Li L, Shimada T, Takahashi H, Ueda H, Fukao Y, Kondo M, Nishimura M, Hara-Nishimura I (2006) MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. The Plant Cell 18, 3535–3547.
| MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhvVKru74%3D&md5=f3e8b58e11b3ba322acfee85c42dc3f9CAS | 17194767PubMed |
Li D-F, Jiang P, Zhu D-Y, Hu Y, Max M, Wang D-C (2008) Crystal structure of Mabinlin II: a novel structural type of sweet proteins and the main structural basis for its sweetness. Journal of Structural Biology 162, 50–62.
| Crystal structure of Mabinlin II: a novel structural type of sweet proteins and the main structural basis for its sweetness.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXksleju7k%3D&md5=4193906a986ca8e3085991b9bf1accb3CAS | 18308584PubMed |
Li L, Shimada T, Takahashi H, Koumoto Y, Shirakawa M, Takagi J, Zhao X, Tu B, Jin H, Shen Z, Han B, Jia M, Kondo M, Nishimura M, Hara-Nishimura I (2013) MAG2 and three MAG2-INTERACTING PROTEINs form an ER-localized complex to facilitate storage protein transport in Arabidopsis thaliana. The Plant Journal 76, 781–791.
| MAG2 and three MAG2-INTERACTING PROTEINs form an ER-localized complex to facilitate storage protein transport in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvVGgsrvN&md5=ce7cf0c234a9ae185570c2b6ca534fdeCAS | 24118572PubMed |
Liu X, Maeda S, Hu Z, Aiuchi T, Nakaya K, Kurihara Y (1993) Purification, complete amino acid sequence and structural characterization of the heat-stable sweet protein, mabinlin II. European Journal of Biochemistry 211, 281–287.
| Purification, complete amino acid sequence and structural characterization of the heat-stable sweet protein, mabinlin II.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXltFWmtrc%3D&md5=e16f3909ee9fda77681bd5cd27b15cfaCAS | 8425538PubMed |
Luckett S, Garcia RS, Barker JJ, Konarev AV, Shewry PR, Clarke AR, Brady RL (1999) High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. Journal of Molecular Biology 290, 525–533.
| High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXktFehsb0%3D&md5=61ff80e5f1c6a1a284a91526c7e5e8c7CAS | 10390350PubMed |
Min W, Jones DH (1994) In vitro splicing of concanavalin A is catalyzed by asparaginyl endopeptidase. Nature Structural & Molecular Biology 1, 502–504.
| In vitro splicing of concanavalin A is catalyzed by asparaginyl endopeptidase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXitVGhs70%3D&md5=a6ee57b1075a2d4b54953a8a47af1686CAS |
Moreno FJ, Clemente A (2008) 2S albumin storage proteins: what makes them food allergens? Open Biochemistry Journal 2, 16–28.
| 2S albumin storage proteins: what makes them food allergens?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXlsleht78%3D&md5=a5290e95b6bd5ab69e93baec2a935e6bCAS | 18949071PubMed |
Mylne JS, Colgrave ML, Daly NL, Chanson AH, Elliott AG, McCallum EJ, Jones A, Craik DJ (2011) Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nature Chemical Biology 7, 257–259.
| Albumins and their processing machinery are hijacked for cyclic peptides in sunflower.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjsVKrsrs%3D&md5=8f74c2813832dee47884851edb84752cCAS | 21423169PubMed |
Mylne JS, Chan LY, Chanson AH, Daly NL, Schaefer H, Bailey TL, Nguyencong P, Cascales L, Craik DJ (2012) Cyclic peptides arising by evolutionary parallelism via asparaginyl-endopeptidase-mediated biosynthesis. The Plant Cell 24, 2765–2778.
| Cyclic peptides arising by evolutionary parallelism via asparaginyl-endopeptidase-mediated biosynthesis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtlChsb%2FN&md5=5b8cb09e5790e6fd4794bd10837ef153CAS | 22822203PubMed |
Onda Y, Nagamine A, Sakurai M, Kumamaru T, Ogawa M, Kawagoe Y (2011) Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. The Plant Cell 23, 210–223.
| Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXpsFaluro%3D&md5=6e362b96158c207879550233a7474409CAS | 21278127PubMed |
Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. The Plant Cell 18, 2567–2581.
| The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1ejurvO&md5=c5063ff25239a342ba9e005aa03ec68dCAS | 17012602PubMed |
Paetzel M, Karla A, Strynadka NCJ, Dalbey RE (2002) Signal peptidases. Chemical Reviews 102, 4549–4580.
| Signal peptidases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XosFSgu7k%3D&md5=0c586814e9b76a074664ff0f0a011009CAS | 12475201PubMed |
Pantoja-Uceda D, Bruix M, Giménez-Gallego G, Rico M, Santoro J (2003) Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis. Biochemistry 42, 13 839–13 847.
| Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXoslKrtLs%3D&md5=8c2d321ba224475928242f84051856f2CAS |
Pantoja-Uceda D, Palomares O, Bruix M, Villalba M, Rodríguez R, Rico M, Santoro J (2004a) Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties. Biochemistry 43, 16 036–16 045.
| Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVWhsbrO&md5=26ee30b42846dc4ef55ca6711c6ff344CAS |
Pantoja-Uceda D, Shewry PR, Bruix M, Tatham AS, Santoro J, Rico M (2004b) Solution structure of a methionine-rich 2S albumin from sunflower seeds: relationship to its allergenic and emulsifying properties. Biochemistry 43, 6976–6986.
| Solution structure of a methionine-rich 2S albumin from sunflower seeds: relationship to its allergenic and emulsifying properties.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjslOitbs%3D&md5=3a5c41e4f6503e6365d72928692516b6CAS | 15170335PubMed |
Rico M, Bruix M, González C, Monsalve RI, Rodríguez R (1996) 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein. Biochemistry 35, 15 672–15 682.
| 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XmvVOisbo%3D&md5=3ff0df4c7b04632047a4ec7e5912d74dCAS |
Rundqvist L, Tengel T, Zdunek J, Björn E, Schleucher J, Alcocer MJC, Larsson G (2012) Solution structure, copper binding and backbone dynamics of recombinant Ber e 1 – the major allergen from brazil nut. PLoS ONE 7, e46435
| Solution structure, copper binding and backbone dynamics of recombinant Ber e 1 – the major allergen from brazil nut.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsFalu7fL&md5=3b3893844e6f12b999b40d261eaeca9dCAS | 23056307PubMed |
Schilling S, Wasternack C, Demuth H-U (2008) Glutaminyl cyclases from animals and plants: a case of functionally convergent protein evolution. Biological Chemistry 389, 983–991.
| Glutaminyl cyclases from animals and plants: a case of functionally convergent protein evolution.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXps1Ohtbc%3D&md5=fe06a35e748c7abea0564b18ba831073CAS | 18979624PubMed |
Sharief FS, Li SS-L (1982) Amino acid sequence of small and large subunits of seed storage protein from Ricinus communis. Journal of Biological Chemistry 257, 14 753–14 759.
Shewry P, Pandya M (1999) The 2S albumin storage proteins. In ‘Seed proteins’. (Eds P Shewry, R Casey) pp. 563–586. (Kluwer: Dordrecht, The Netherlands)
Shewry PR, Napier JA, Tatham AS (1995) Seed storage proteins: structures and biosynthesis. The Plant Cell 7, 945–956.
Shimada T, Kuroyanagi M, Nishimura M, Hara-Nishimura I (1997) A pumpkin 72-kDa membrane protein of precursor-accumulating vesicles has characteristics of a vacuolar sorting receptor. Plant & Cell Physiology 38, 1414–1420.
| A pumpkin 72-kDa membrane protein of precursor-accumulating vesicles has characteristics of a vacuolar sorting receptor.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXlsVem&md5=279dfae7c78a68d04828267df84b25b3CAS |
Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I (2003a) Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 100, 16 095–16 100.
| Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVGjtQ%3D%3D&md5=a97e114931ad333f72874305ce30c9e8CAS |
Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, Kobayashi M, Kondo M, Nishimura M, Hara-Nishimura I (2003b) Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. Journal of Biological Chemistry 278, 32 292–32 299.
| Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXmsVOmsbk%3D&md5=2f986a0b57b41798662af944d5a9a439CAS |
Shimada T, Koumoto Y, Li L, Yamazaki M, Kondo M, Nishimura M, Hara-Nishimura I (2006) AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant & Cell Physiology 47, 1187–1194.
| AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFCgur7L&md5=1d3bc4015a575d0df1bb7c3ae71d6a54CAS |
Takagi J, Renna L, Takahashi H, Koumoto Y, Tamura K, Stefano G, Fukao Y, Kondo M, Nishimura M, Shimada T, Brandizzi F, Hara-Nishimura I (2013) MAIGO5 functions in protein export from Golgi-associated endoplasmic reticulum exit sites in Arabidopsis. The Plant Cell 25, 4658–4675.
| MAIGO5 functions in protein export from Golgi-associated endoplasmic reticulum exit sites in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXitVOqsrs%3D&md5=c5c8f7a84bebb20aa8567f89e14ce753CAS | 24280388PubMed |
Takahashi H, Tamura K, Takagi J, Koumoto Y, Hara-Nishimura I, Shimada T (2010) MAG4/Atp115 is a Golgi-localized tethering factor that mediates efficient anterograde transport in Arabidopsis. Plant & Cell Physiology 51, 1777–1787.
| MAG4/Atp115 is a Golgi-localized tethering factor that mediates efficient anterograde transport in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXht1OgsrfK&md5=8bd936b5cb7a91794a8f361c4330ff18CAS |
Thoyts PJE, Napier JA, Millichip M, Stobart AK, Griffiths WT, Tatham AS, Shewry PR (1996) Characterization of a sunflower seed albumin which associates with oil bodies. Plant Science 118, 119–125.
| Characterization of a sunflower seed albumin which associates with oil bodies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XktlSitbg%3D&md5=7cd6ec1a105e95a6047f034c56a09c0eCAS |
Yamada K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I (1999) Multiple functional proteins are produced by cleaving Asn-Gln bonds of a single precursor by vacuolar processing enzyme. Journal of Biological Chemistry 274, 2563–2570.
| Multiple functional proteins are produced by cleaving Asn-Gln bonds of a single precursor by vacuolar processing enzyme.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXovVKjsQ%3D%3D&md5=2eb07b620a30ba63fe59d7ce1a3d43baCAS | 9891029PubMed |
Yamazaki M, Shimada T, Takahashi H, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I (2008) Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant & Cell Physiology 49, 142–156.
| Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXjsVWltbc%3D&md5=be8dafabf09694579e91531a1323690dCAS |