DNA barcodes and morphology reveal a hybrid hawkmoth in Tahiti (Lepidoptera : Sphingidae)
R. Rougerie A E , Jean Haxaire B , Ian J. Kitching C and Paul D. N. Hebert DA Laboratoire d’Ecologie, EA 1293 ECODIV, UFR Sciences et Techniques, Université de Rouen, 76821 Mont Saint Aignan Cedex, France.
B Honorary Attaché, Muséum national d’Histoire naturelle de Paris, Le Roc, F-47310 Laplume, France.
C Department of Life Sciences, Natural History Museum, London, SW7 5BD, UK.
D Department of Integrative Biology & Biodiversity Institute of Ontario, University of Guelph, Guelph, ON, N1G 2W1, Canada.
E Corresponding author. Email: rrougeri@gmail.com
Invertebrate Systematics 26(6) 445-450 https://doi.org/10.1071/IS12029
Submitted: 17 April 2012 Accepted: 13 September 2012 Published: 19 December 2012
Journal Compilation © CSIRO Publishing 2012 Open Access CC BY-NC-ND
Abstract
Interspecific hybridisation is a rare but widespread phenomenon identified as a potential complicating factor for the identification of species through DNA barcoding. Hybrids can, however, also deceive morphology-based taxonomy, resulting in the description of invalid species based on hybrid specimens. As the result of an unexpected case of discordance between barcoding results and current morphology-based taxonomy, we discovered an example of such a hybrid ‘species’ in hawkmoths. By combining barcodes, morphology and a nuclear marker, we show that Gnathothlibus collardi Haxaire, 2002 is actually an F1 hybrid between two closely related species that co-occur on Tahiti. In accordance with the International Code of Zoological Nomenclature, the taxon G. collardi is thus invalid as a species. This study demonstrates the potential of DNA barcodes to detect overlooked hybrid taxa. With the growth of sequence libraries, we anticipate that more unsuspected hybrid species will be detected, particularly among those taxa that are very rare, such as those known from only the type specimen.
Additional keywords: 28S rDNA, COI, hybrid detection.
Introduction
Natural interspecific hybridisation is a rare but widespread phenomenon in animal taxa (Mayr 1942; Seehausen 2004; Mallet 2008; Schwenk et al. 2008). Because it can lead to introgression between species, hybridisation can complicate the use of DNA-based approaches for species identification such as DNA barcoding (Vences et al. 2005; Whitworth et al. 2007; Schmidt and Sperling 2008) by causing discordance between the species-tree and the gene-tree in which species appear para- or polyphyletic (Funk and Omland 2003; Bachtrog et al. 2006; Zakharov et al. 2009). However, the complicating effects of hybridisation are not restricted to DNA-based identification methods; they can also strongly affect morphological identification (Thulin et al. 2006). Between closely related species that lack clear-cut diagnostic characters, hybrids can be confounded with intraspecific variation in one or other parental species. Undetected, they may blur the delineation of diagnostic characters between the two hybridising species. Alternatively, hybrids can be different enough from both parent species to gain description as a distinct species. Such cases have seldom been investigated in animals (but see Graves (1990) and Parham et al. (2001)), though they represent erroneous species accounts with a potential impact on taxonomy users. Resources may, for instance, be wasted on conservation of what are thought to be rare and endangered species, but which are actually hybrids (Allendorf et al. 2001). Interestingly, in cases where species might have been described erroneously on the basis of hybrid specimens, the discordance between morphological diagnostic characters and mtDNA offers a way of detecting a possible hybrid specimen, identifying its maternal parent species, and then triggering further study to confirm its status.
In this paper, we present a case study in which the investigation of a case of discordance between DNA barcodes and morphology revealed that Gnathothlibus collardi Haxaire, 2002, a rare hawkmoth species (Haxaire 2002), was described from specimens resulting from the hybridisation of two species on Tahiti. We present morphological and nuclear DNA evidence supporting this conclusion.
Materials and methods
Specimen sampling
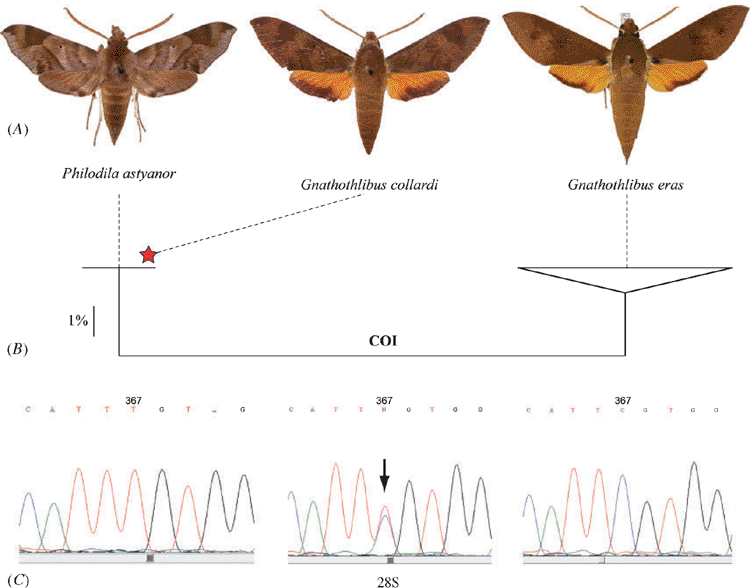
In the course of the global DNA barcoding campaign for sphingid moths (or hawkmoths), specimens of Philodila astyanor (Boisduval, [1875]), Gnathothlibus collardi, and G. eras (Boisduval, 1832) were sampled from Tahiti, the largest of the Society Islands in French Polynesia. Whereas the first two species are endemic to Tahiti, the third has a broad range extending through Melanesia to Sulawesi and the northern and eastern coasts of Australia. Samples analysed for this species include representatives from Tahiti, Bora Bora, Australia (Northern Territory, Queensland and Dauan Island) and Sulawesi. In total, 22 specimens were sampled for DNA barcoding and included in this study: 2 paratypes of G. collardi, 4 specimens of P. astyanor, and 16 specimens of G. eras (4 of these from Tahiti). Details of the specimens included in this study are given in Table 1; each record is given a unique specimen identifier (SampleID) and sequence identifier (ProcessID). Collection data, a photograph and ancillary information, such as the collection holding each specimen, are available in BOLD, the Barcode of Life Datasystems (Ratnasingham and Hebert 2007) in public projects.
|
DNA extraction, sequencing and data accessibility
The protocol for DNA extraction, amplification and sequencing followed the standard high-throughput procedure developed and used for Lepidoptera at the Canadian Centre for DNA Barcoding (see Vaglia et al. (2008) for details). This procedure includes a multistep iterative PCR strategy in which DNA samples that fail to amplify are ‘hit-picked’ and go through additional PCR amplification attempts targeting shorter amplicons (Decaëns and Rougerie 2008). After we realised that Gnathothlibus collardi specimens might be hybrids between G. eras and Philodila astyanor, we sought additional evidence by sequencing a nuclear marker. From the same DNA extracts used to generate the DNA barcode sequences, we amplified and sequenced the D2 expansion segment of the large ribosomal subunit (28S). This fragment was amplified using the primer pair D2B and D3Ar (Saux et al. 2004). The PCR cocktail was similar to that used for COI, but a different thermocycling regime was employed: 1 cycle of 2 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at 56°C, and 2 min at 72°C, with a final step of 2 min at 72°C. Contigs were assembled using SeqScape ver. 2.1.1 (Applied Biosystems, Carlsbad, CA) and Sequencher 4.5 (Gene Code Corporation, Ann Arbor, MI), and were subsequently aligned using Bioedit ver. 7.0.5.3 (Hall 1999). The absence of indels in both the COI and 28S sequences made sequence alignment straightforward and unambiguous. Along with specimen data, sequence data for both genes are available on BOLD within public projects and in the assembled dataset SPHhyb01. All sequences are also available on GenBank with the accession numbers listed in Table 1.
Sequence analyses
The consistency of DNA barcodes as diagnostic molecular markers for the three taxa studied was tested through sequence similarity analysis, using the Management and Analysis component of BOLD (Ratnasingham and Hebert 2007) to compute K2P distances as a measure of intra- and interspecific variation. Neighbour-Joining (NJ) analysis, as implemented in BOLD, was used to construct the distance tree. Diagnostic characters in 28S rDNA sequences were searched for visually using Bioedit.
Morphological examination
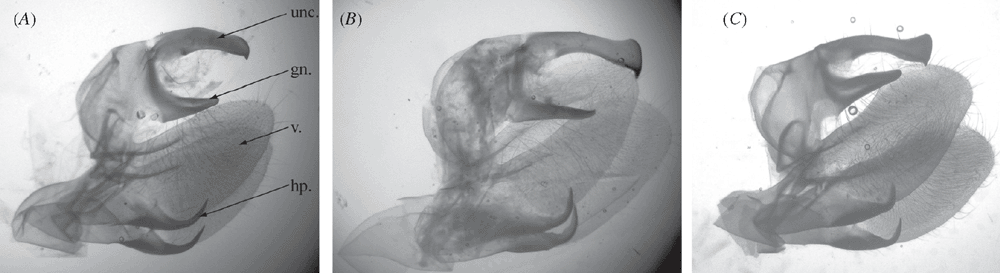
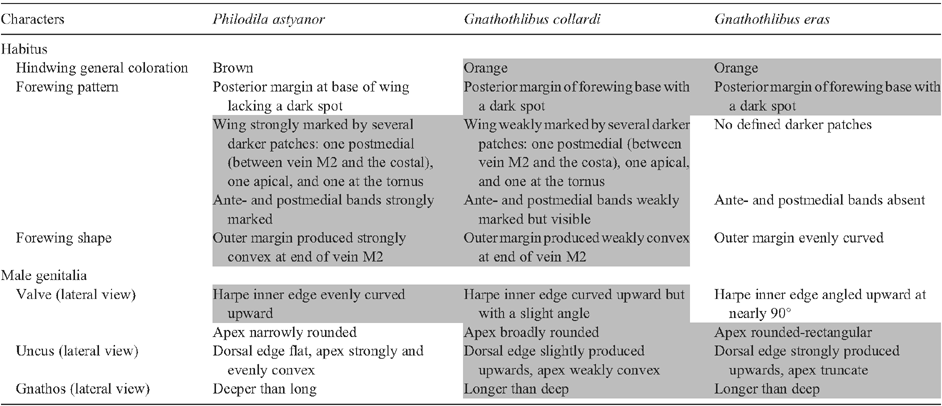
We examined several dozen specimens of Gnathothlibus eras, either directly in collections or as photographs, three male specimens of G. collardi including the holotype and two paratypes (to our knowledge, there is only one other specimen known for that species), and 37 specimens of Philodila astyanor, including a photograph of the holotype preserved at the Carnegie Museum of Natural History (CMNH, Pittsburgh, USA). The male genitalia of the three taxa were prepared using standard techniques and also compared.
Results
DNA barcoding
The COI region used as the standard DNA barcode in animals was recovered in all samples except for one of the paratypes of Gnathothlibus collardi, the age of which (>20 years old) probably prevented amplification of the target sequence despite several attempts to amplify shorter fragments (see Material and methods). In total, 21 barcodes were recovered representing four specimens of Philodila astyanor, 16 of G. eras and one of G. collardi. Sequence similarity analysis using K2P distances revealed a clear discrimination between the first two species, each of which formed a cohesive genetic unit as illustrated in the NJ tree (Fig. 1B). The minimum sequence divergence observed between the two species is 4.99%; all four specimens of P. astyanor possessed an identical DNA barcode, while G. eras specimens show some intraspecific variation (maximum and mean intraspecific distance of 0.83% and 0.35% (s.e. = 0.023%, n = 16) respectively). The presence of a clear barcode divergence was not surprising given the obvious morphological differences between these two species (Fig. 1A). However, our analysis unexpectedly revealed that the paratype of G. collardi possessed a barcode sequence identical to that of P. astyanor (Fig. 1B) despite the morphological dissimilarity between the two taxa (G. collardi resembles G. eras at first glance (Fig. 1A), and its male genitalia are also more similar to those of G. eras (Figs 2B, C, compared with 2A)). Cross-contamination between the two species was ruled out by reprocessing a second tissue sample from the same G. collardi specimen, which yielded identical results.
|
Test of the hybrid hypothesis
The barcode congruence between the specimen of G. collardi and P. astyanor could be explained if G. collardi specimens are F1 hybrids between female P. astyanor and male G. eras. This hybrid hypothesis would explain the morphological distinctiveness of G. collardi as well as its shared DNA barcode sequence through maternal inheritance of the mitochondrial genome. Examination of the D2 region of the 28S rDNA gene – a nuclear gene inherited from both parents – from three specimens each of the hypothetical parent species, revealed no variation except for a single diagnostic substitution distinguishing P. astyanor and G. eras. The two species showed a consistent difference at Position 367 of the amplified fragment of 28S (Fig. 1C), with a thymine in P. astyanor versus a cytosine in G. eras. In G. collardi, this position is heterozygous, as electropherograms show a cytosine + thymine double peak (Fig. 1C), supporting the hybrid hypothesis. A more thorough morphological reexamination of specimens of the three species then revealed that G. collardi displays a mosaic of characters found in its parent species (Table 2; Figs 1A, 2A–C). Because the species name, Gnathothlibus collardi, is based on a type specimen of hybrid origin, it is invalid (ICZN 1999, Article 23.8: ‘A species-group name established for an animal later found to be a hybrid must not be used as the valid name for either of the parental species’).
|
Discussion
DNA barcodes revealed an unexpected case of a species invalidly described from hybrid specimens. The initial discordance between morphological identifications and the cohesiveness of DNA barcode clusters provoked deeper investigation of the situation with both a nuclear marker and morphology. These analyses showed that natural hybrid specimens between Philodila astyanor and G. eras in Tahiti had deceived a taxonomist into describing an invalid species. This represents the second known example in the Sphingidae as a similar case was recently reported by Hundsdoerfer et al. (2011), who inferred a hybrid origin for Hyles sammuti Eitschberger, Danner & Surholt 1998 from the discordance between morphology and mtDNA sequence data. However, they suggested retaining the name sammuti for convenience while awaiting confirmation from nuclear DNA sequences.
Although it is difficult to estimate how frequently taxonomists have erroneously described species from hybrids, we note that DNA barcode libraries assembled in collaboration with expert taxonomists can efficiently reveal cases where the interspecific hybrids differ phenotypically from their parent species. Rare species, such as those known only from the type specimen, may well involve a higher incidence of hybrid taxa. This status could explain why some species elude modern re-collection and are thought to be extinct. For instance, the African hawkmoth Hippotion chloris Rothschild & Jordan, 1907 is known only from the holotype, and Carcasson (1968) suggested that it was a hybrid between Hippotion celerio (Linnaeus, 1758) and Basiothia medea (Fabricius, 1781) because of its intermediate appearance between these taxa. Barcode records reveal that the two hypothetical parent species are more than 4% divergent (Wilson et al. 2011), with nearest neighbours respectively at 2.3% (Hippotion aurora Rothschild & Jordan, 1903) and 3.3% (Basiothia laticornis (Butler, 1879)). As a result, the recovery of even a short fragment of the barcode region from the holotype of H. chloris, as has already been achieved successfully for archival material in Lepidoptera (Hausmann et al. 2009a, 2009b), would allow for quick support or refutation of the hybrid hypothesis. In hawkmoths, rarity and intermediate phenotypes suggest that a similar hybrid status may also be hypothesised for Xylophanes clarki Ramsden, 1921, a possible hybrid between X. tersa (Linnaeus, 1771) and X. pluto (Fabricius, 1777), and Hyles churkini Saldaitis & Ivinskis, 2006, a possible hybrid between H. zygophylli (Ochsenheimer, 1808) and H. euphorbiae (Linnaeus, 1758) or H. costata (von Nordmann, 1851).
A high incidence of hybridisation has been reported in some groups of Lepidoptera, ranging from 6–15% within the genus Papilio (Sperling 1990) to as high as 29% within the tribe Heliconiina (Mallet et al. 2007). These reports suggest that F1 hybrids may account for several species, especially in groups like butterflies, hawkmoths and silkmoths where a significant fraction of species have been described from one or a few specimens. Taxonomic ‘exaggeration’ (Pillon and Chase 2007) caused by hybrids is certainly known from other groups that have seen intensive taxonomic study, including hummingbirds (Graves 1990) and turtles (Stuart and Parham 2007). Given the significant proportion of species described upon a single specimen, one in six newly described species of invertebrates during the past decade, and one in four for vertebrates (Lim et al. 2012), it is worth testing to determine how many were based on hybrid specimens.
DNA barcode reference libraries assembled with the involvement of expert taxonomists can help to reveal the impact of hybridisation on taxonomy, limiting the consequences of ‘bad taxonomy’ (Bortolus 2008). With possibly increasing rates of hybridisation and introgression as the result of species-range alteration by human activities (through modifications of habitats, translocations of organisms, climatic changes, etc.), interspecific hybrids may cause increasing taxonomic confusion. Importantly, these hybrid specimens or populations may have important implications for conservation (Brownlow 1996; Allendorf et al. 2001; vonHoldt et al. 2011) and their early detection can be critical to preventing local or even global extinction of parent species (‘speciation reversal’, see e.g. Vonlanthen et al. (2012)). In this respect, DNA barcode libraries may act as sentinels for the early detection of hybridisation.
Acknowledgements
We are particularly grateful to staff at the Canadian Centre for DNA Barcoding (University of Guelph, Canada) for their help in processing the specimens examined in this study. We thank Dr John Rawlins (Carnegie Museum of Natural History), who hosted IJK during his visits to the CMNH in 1992 and 1998, as well as Dr John Lasalle (Australian National Insect Collection, Canberra), Steeve Collard, Dominique Bernaud, Yves Estradel and Georges Orhant for contributing material in their collections or for sharing specimens they collected. Special credit is due to Steeve Collard who suggested the hybrid status when he reported the capture of the first specimen of Gnathothlibus collardi to JH; he abandoned this notion after collecting the third specimen on the basis that hybrids are exceptional in nature. Funding was provided by the government of Canada through Genome Canada and the Ontario Genomics Institute in support of the International Barcode of Life project, and by NSERC.
References
Allendorf, F. W., Leary, R. F., Spruell, P., and Wenburg, J. K. (2001). The problems with hybrids: setting conservation guidelines. Trends in Ecology & Evolution 16, 613–622.| The problems with hybrids: setting conservation guidelines.Crossref | GoogleScholarGoogle Scholar |
Bachtrog, D., Thornton, K., Clark, A., and Andolfatto, P. (2006). Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution 60, 292–302.
| 1:CAS:528:DC%2BD28XjsFaqt7c%3D&md5=032518e1536961b7d3f5da11da35fa3cCAS |
Bortolus, A. (2008). Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology. Ambio 37, 114–118.
| Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology.Crossref | GoogleScholarGoogle Scholar |
Brownlow, C. A. (1996). Molecular taxonomy and the conservation of the red wolf and other endangered carnivores. Conservation Biology 10, 390–396.
| Molecular taxonomy and the conservation of the red wolf and other endangered carnivores.Crossref | GoogleScholarGoogle Scholar |
Carcasson, R. H. (1968). Revised catalogue of the African Sphingidae (Lepidoptera) with descriptions of the East African species. Journal of the East Africa Natural History Society and National Museum 26, 1–148.
Decaëns, T., and Rougerie, R. (2008). Descriptions of two new species of Hemileucinae (Lepidoptera: Saturniidae) from the region of Muzo in Colombia – evidence from morphology and DNA barcodes. Zootaxa 1944, 34–52.
Funk, D. J., and Omland, K. E. (2003). Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology Evolution and Systematics 34, 397–423.
| Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA.Crossref | GoogleScholarGoogle Scholar |
Graves, G. (1990). Systematics of the “green-throated sunangels” (Aves: Trochilidae): valid taxa or hybrids? Proceedings of the Biological Society of Washington 103, 6–25.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98.
| 1:CAS:528:DC%2BD3cXhtVyjs7Y%3D&md5=08d747d827542961412adcc224832fc5CAS |
Hausmann, A., Hebert, P. D. N., Mitchell, A., Rougerie, R., Sommerer, M., Edwards, T., and Young, C. J. (2009a). Revision of the Australian Oenochroma vinaria Guenee, 1858 species-complex (Lepidoptera: Geometridae, Oenochrominae): DNA barcoding reveals cryptic diversity and assesses status of type specimen without dissection. Zootaxa 2239, 1–21.
Hausmann, A., Sommerer, M., Rougerie, R., and Hebert, P. (2009b). Hypobapta tachyhalotaria spec. nov from Tasmania – an example of a new species revealed by DNA barcoding. Spixiana 32, 161–166.
Haxaire, J. (2002). Description d’un nouveau Sphingidae de l’île de Tahiti: Gnathothlibus collardi (Lepidoptera: Sphingidae). Lambillionea 102, 495–499.
Hundsdoerfer, A. K., Mende, M. B., Kitching, I. J., and Cordellier, M. (2011). Taxonomy, phylogeography and climate relations of the Western Palaearctic spurge hawkmoth (Lepidoptera, Sphingidae, Macroglossinae). Zoologica Scripta 40, 403–417.
| Taxonomy, phylogeography and climate relations of the Western Palaearctic spurge hawkmoth (Lepidoptera, Sphingidae, Macroglossinae).Crossref | GoogleScholarGoogle Scholar |
ICZN (1999). ‘International Code of Zoological Nomenclature.’ 4th edn. (The International Trust for Zoological Nomenclature: London, UK.)
Lim, G. S., Balke, M., and Meier, R. (2012). Determining species boundaries in a world full of rarity: singletons, species delimitation methods. Systematic Biology 61, 165–169.
| Determining species boundaries in a world full of rarity: singletons, species delimitation methods.Crossref | GoogleScholarGoogle Scholar |
Mallet, J. (2008). Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philosophical Transactions of the Royal Society B. Biological Sciences 363, 2971–2986.
| Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation.Crossref | GoogleScholarGoogle Scholar |
Mallet, J., Beltran, M., Neukirchen, W., and Linares, M. (2007). Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evolutionary Biology 7, 28.
| Natural hybridization in heliconiine butterflies: the species boundary as a continuum.Crossref | GoogleScholarGoogle Scholar |
Mayr, E. (Ed.) (1942). ‘Systematics and Origin of Species.’ (Columbia University Press: New York.)
Parham, J. F., Simison, W. B., Kozak, K. H., Feldman, C. R., and Shi, H. (2001). New Chinese turtles: endangered or invalid? A reassessment of two species using mitochondrial DNA, allozyme electrophoresis and known-locality specimens. Animal Conservation 4, 357–367.
| New Chinese turtles: endangered or invalid? A reassessment of two species using mitochondrial DNA, allozyme electrophoresis and known-locality specimens.Crossref | GoogleScholarGoogle Scholar |
Pillon, Y., and Chase, M. W. (2007). Taxonomic exaggeration and its effects on orchid conservation. Conservation Biology 21, 263–265.
| Taxonomic exaggeration and its effects on orchid conservation.Crossref | GoogleScholarGoogle Scholar |
Ratnasingham, S., and Hebert, P. D. N. (2007). BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Molecular Ecology Notes 7, 355–364.
| BOLD: The Barcode of Life Data System (http://www.barcodinglife.org).Crossref | http://www.barcodinglife.org).&journal=Molecular Ecology Notes&volume=7&pages=355-364&publication_year=2007&author=S%2E%20Ratnasingham&hl=en&doi=10.1111/j.1471-8286.2007.01678.x" target="_blank" rel="nofollow noopener noreferrer" class="reftools">GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXntVyksbc%3D&md5=53cd40598900a88b3cc1c91ad7b9427cCAS |
Saux, C., Fisher, B. L., and Spicer, G. S. (2004). Dracula ant phylogeny as inferred by nuclear 28S rDNA sequences and implications for ant systematics (Hymenoptera: Formicidae: Amblyoponinae). Molecular Phylogenetics and Evolution 33, 457–468.
| Dracula ant phylogeny as inferred by nuclear 28S rDNA sequences and implications for ant systematics (Hymenoptera: Formicidae: Amblyoponinae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXntVKqu7g%3D&md5=90c470efe034e5e06033fbdc1ae3c1d5CAS |
Schmidt, B. C., and Sperling, F. A. (2008). Widespread decoupling of mtDNA variation and species integrity in Grammia tiger moths (Lepidoptera: Noctuidae). Systematic Entomology 33, 613–634.
| Widespread decoupling of mtDNA variation and species integrity in Grammia tiger moths (Lepidoptera: Noctuidae).Crossref | GoogleScholarGoogle Scholar |
Schwenk, K., Brede, N., and Streit, B. (2008). Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Philosophical Transactions of the Royal Society B. Biological Sciences 363, 2805–2811.
| Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals.Crossref | GoogleScholarGoogle Scholar |
Seehausen, O. (2004). Hybridization and adaptive radiation. Trends in Ecology & Evolution 19, 198–207.
| Hybridization and adaptive radiation.Crossref | GoogleScholarGoogle Scholar |
Sperling, F. A. H. (1990). Natural hybrids of Papilio (Insecta: Lepidoptera): poor taxonomy or interesting evolutionary process? Canadian Journal of Zoology 68, 1790–1799.
| Natural hybrids of Papilio (Insecta: Lepidoptera): poor taxonomy or interesting evolutionary process?Crossref | GoogleScholarGoogle Scholar |
Stuart, B. L., and Parham, J. F. (2007). Recent hybrid origin of three rare Chinese turtles. Conservation Genetics 8, 169–175.
| Recent hybrid origin of three rare Chinese turtles.Crossref | GoogleScholarGoogle Scholar |
Thulin, C.-G., Stone, J., Tegelström, H., and Walker, C. W. (2006). Species assignment and identification among Scandinavian hares Lepus europaeus and L. timidus. Wildlife Biology 12, 29–38.
| Species assignment and identification among Scandinavian hares Lepus europaeus and L. timidus.Crossref | GoogleScholarGoogle Scholar |
Vaglia, T., Haxaire, J., Kitching, I. J., Meusnier, I., and Rougerie, R. (2008). Morphology and DNA barcoding reveal three cryptic species within the Xylophanes neoptolemus and loelia species-groups (Lepidoptera: Sphingidae). Zootaxa 1923, 18–36.
Vences, M., Thomas, M., Bonett, R. M., and Vieites, D. R. (2005). Deciphering amphibian diversity through DNA barcoding: chances and challenges. Philosophical Transactions of the Royal Society B. Biological Sciences 360, 1859–1868.
| Deciphering amphibian diversity through DNA barcoding: chances and challenges.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtlSjsrjE&md5=2eec6fc25c94b8fa4ec087fcdffdb959CAS |
vonHoldt, B. M., Pollinger, J. P., Earl, D. A., Knowles, J. C., Boyko, A. R., Parker, H., Geffen, E., Pilot, M., Jedrzejewski, W., Jedrzejewska, B., Sidorovich, V., Greco, C., Randi, E., Musiani, M., Kays, R., Bustamante, C. D., Ostrander, E. A., Novembre, J., and Wayne, R. K. (2011). A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Research 21, 1294–1305.
| A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXpvFeqs7k%3D&md5=a8697e6ff407d38139859011c514efe9CAS |
Vonlanthen, P., Bittner, D., Hudson, A. G., Young, K. A., Muller, R., Lundsgaard-Hansen, B., Roy, D., Di Piazza, S., Largiader, C. R., and Seehausen, O. (2012). Eutrophication causes speciation reversal in whitefish adaptive radiations. Nature 482, 357–362.
| Eutrophication causes speciation reversal in whitefish adaptive radiations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xit12gtbY%3D&md5=6c2651df9f02de8d0f6b46d71e1000c2CAS |
Whitworth, T. L., Dawson, R. D., Magalon, H., and Baudry, E. (2007). DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). Proceedings. Biological Sciences 274, 1731–1739.
| DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXosVans7k%3D&md5=78b3201e91b8dfa011fecf87056696bcCAS |
Wilson, J. J., Rougerie, R., Schonfeld, J., Janzen, D. H., Hallwachs, W., Hajibabaei, M., Kitching, I. J., Haxaire, J., and Hebert, P. D. N. (2011). When species matches are unavailable are DNA barcodes correctly assigned to higher taxa? An assessment using sphingid moths. BMC Ecology 11, 18.
| When species matches are unavailable are DNA barcodes correctly assigned to higher taxa? An assessment using sphingid moths.Crossref | GoogleScholarGoogle Scholar |
Zakharov, E. V., Lobo, N. F., Nowak, C., and Hellmann, J. J. (2009). Introgression as a likely cause of mtDNA paraphyly in two allopatric skippers (Lepidoptera: Hesperiidae). Heredity 102, 590–599.
| Introgression as a likely cause of mtDNA paraphyly in two allopatric skippers (Lepidoptera: Hesperiidae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmtFSisr0%3D&md5=e24f2ba59e1d83eb68ffc23b96d9aa55CAS |