Using MLVA to type strains of Salmonella Typhimurium in New South Wales
Gwendolyn L. GilbertCentre for Infectious Diseases and Microbiology, Institute of Clinical Pathology and Medical Research, Westmead Hospital, Westmead. Email: l.gilbert@usyd.edu.au
NSW Public Health Bulletin 19(2) 29-31 https://doi.org/10.1071/NB07116
Published: 28 February 2008
Abstract
Summary: Phage typing has been the traditional strain typing (or ‘fingerprinting’) method used in Australia for surveillance of common salmonella serovars (such as Salmonella Typhimurium) and outbreak investigations. The need for more accessible, discriminatory and objective methods has been recognised but, until now, none has been widely accepted. Recently, the molecular typing method, known as MLVA (multilocus variable number tandem repeat analysis), has been applied to several Salmonella serovars and promises to provide faster strain typing and cluster identification than phage typing, with comparable or better sensitivity. The present article is intended as a short primer on MLVA typing, which has recently been introduced into routine use at the New South Wales Enteric Reference Laboratory at the Centre for Infectious Diseases and Microbiology, Institute of Clincial Pathology and Medical Research, Westmead.
The New South Wales Enteric Reference Laboratory receives 1800–2000 Salmonella isolates for serotyping each year, of which more than 50 per cent are S. Typhimurium. Such large numbers of a single serotype make it very difficult to identify potential outbreaks without additional strain typing (or ‘fingerprinting’). Phage typing has provided valuable epidemiological data and assisted in outbreak investigations for nearly 60 years but has major limitations and is increasingly inadequate for 21st century disease surveillance.
Current methods for subtyping Salmonella
Phage typing has been, until recently, the subtyping method of choice for S. Typhimurium and several molecular typing methods are also used when further discrimination is needed. These methods are described briefly in the article by Wang et al. in this issue.
Introducing multi-locus variable number tandem repeat analysis
Recently, multi-locus variable number tandem repeat analysis has been successfully applied to many bacterial species, including several Salmonella serotypes, and has the potential to largely replace both phage typing and pulsed field gel electrophoresis as the primary subtyping method for salmonellae.1–4
Most bacterial genomes contain several sites or loci (genes or intergenic sequences), which contain variable numbers of repeated sequences that may be duplicated or deleted as part of the natural genetic variation of the species. This means that the total length of the locus varies between different strains. Development of a multi-locus variable number tandem repeat analysis (MLVA) scheme for a particular organism involves identifying up to 10 suitable loci within the genome. Suitability depends on the length of each sequence, by how much and how frequently the numbers of sequences vary, and whether there are conserved flanking sequences at each end that can be targeted by polymerase chain reaction primers. Strain-specific profiles derived from examination of these loci, allow objective strain comparison.
MLVA involves first amplifying the target loci by polymerase chain reaction and then measuring (either by gel or capillary electrophoresis) the lengths of the amplified DNA segments (amplicons). The number of repeats for each locus is inferred by subtracting the known length of the flanking sequence from the total amplicon length and dividing the result by the known length of each repeat sequence (as illustrated in Figure 1). The MLVA result or strain-specific profile is a series of numbers, each of which represents the number of repeats at one of the loci in a standard order.
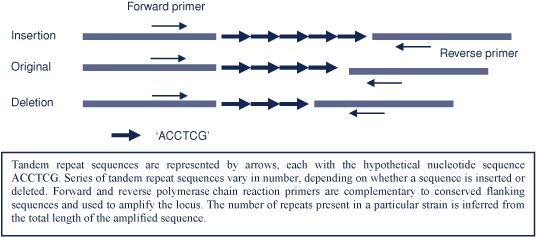
|
For S. Typhimurium, loci are designated as STTR – Salmonella Typhimurium tandem repeat – and an arbitrary number. The scheme devised by Lindstedt et al., involves five loci – STTR9, STTR5, STTR6, STTR10pl (‘pl’ refers to the fact that this locus – STTR10pl – is on a plasmid, whereas the other loci are on the chromosome) and STTR3.2 The lengths of repeat sequences at these loci, in base pairs, are: 9 for STTR9, 6 for STTR5, 6 for STTR6, 6 for STTR10pl and a combination of 27 and 33 base pair repeats for STTR3. There are various possible formats in which the MLVA profile could be expressed but, so far, none has been generally adopted.1,5 Recently, representatives from several Australian reference laboratories agreed on the following convention for S. Typhimurium. For all loci except STTR3, the result will be expressed as 0 if there is no amplicon (i.e. the locus is absent); 1 if the size of the amplicon corresponds with that of the flanking region (i.e. the locus is present but no repeat sequences are present); 2 if the amplicon length corresponds with the sum of the flanking region and one repeat, and so on.
For STTR3, which is complicated by the potential presence of variable numbers of repeats of two different lengths, it was agreed that the actual amplicon length would be given (although it is possible to calculate the actual number of repeats of each length). This agreed convention is illustrated in Figure 2. This coding system may need to be modified in future but, in the meantime, it provides a method by which Australian laboratories can compare results.
A quality assurance program has recently been introduced; the first panel of isolates has been distributed and tested to ensure that the methods followed in participating laboratories generate consistent results. Consistency, is essential to enable the identification of disease outbreaks that cross state borders. A similar successful quality assurance exercise was recently reported from Scandinavia.5
In a recent study of 168 S. Typhimurium isolates, representing 46 phage types, STTR3, STTR5 and STTR9 were present in all isolates tested, STTR6 was present in 96 per cent and STTR10pl in 85 per cent of isolates. The numbers of repeats varied at different loci from as few as one or two for STTR9 to as many as 30 for STTR5 (Wang Q, Kong F, Jelfs P, Gilbert GL, unpublished data).
Using MLVA to identify clusters of disease
An important issue that is yet to be decided is the definition of a cluster. This requires further investigation. Preliminary data show that there is a high rate of clustering of isolates (when a cluster is defined as two or more isolates with the same MLVA profile). For example, during a 4-month period, 85 per cent of 185 S. Typhimurium isolates received consecutively by the NSW Enteric Reference Laboratory and tested by MLVA, were clustered, with 2–20 isolates per cluster. Over a longer period, it is likely that nearly all isolates would be clustered – that is, few, if any, individual MLVA profiles will be unique.
It is impractical to investigate every cluster, irrespective of the frequency or distribution of individual cases. The number that can be investigated will depend on available resources. One proposed cluster definition, suitable for a relatively low incidence country like Australia, is five or more cases of the same MLVA type occurring in a defined geographic area in a 4-week period.6 Using this definition, 59 per cent of the 185 NSW isolates were clustered into 6 clusters over 4 months – a more feasible number for follow-up. Because of the relatively short time period in which a cluster is defined, the chance of identifying a source is relatively high.
Finally we need to determine the level of variation between isolates that can occur before isolates are no longer regarded as belonging to the same outbreak or cluster. The loss or gain of repeats occurs quite frequently at loci 2–4 but rarely at loci 1 and 5. Thus, profiles that vary by one or two digits at one of loci 2–4 can be regarded as probably related and investigated accordingly. Isolates are less likely to be related if there are differences at two of the inner loci, and are very unlikely to be related if there are differences at all three inner loci or at either locus 1 or locus 5. Further experience is required to develop more precise cluster definitions.
Next steps
During the next 12 months, the NSW Enteric Reference Laboratory, in collaboration with the Communicable Diseases Branch of the NSW Department of Health and the NSW Food Authority, will be evaluating MLVA prospectively, by comparing the results available within approximately 2 weeks of the receipt of isolates with those of epidemiological investigations of suspected clusters. We will also evaluate a novel molecular phage type identification system developed in our laboratory, which provides complementary information. In addition, we aim to develop a web-based reporting system. This will describe the geographic distribution of cases and clusters based on postcodes over defined time periods (spatiotemporal distribution) and will assess the risk that an individual case is part of a cluster based on detailed analysis of MLVA data.
Acknowledgements
I would like to thank Qinning Wang, Vitali Sintchenko, Peter Howard, Peter Jelfs and Robert Chiew for their contributions to the work described in the present study. Qinning Wang kindly provided images and data for the figures.
[1] Hyytia-Trees E, Smole SC, Fields PA, Swaminathan B, Ribot EM. Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne Pathog Dis 2006; 3(1): 118–31.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[2] Lindstedt BA, Vardund T, Aas L, Kapperud G. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J Microbiol Methods 2004; 59(2): 163–72.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[3] Ramisse V, Houssu P, Hernandez E, Denoeud F, Hilaire V, Lisanti O, et al. Variable number of tandem repeats in Salmonella enterica subsp. enterica for typing purposes. J Clin Microbiol 2004; 42(12): 5722–30.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[4] Boxrud D, Pederson-Gulrud K, Wotton J, Medus C, Lyszkowicz E, Besser J, et al. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J Clin Microbiol 2007; 45(2): 536–43.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[5] Lindstedt BA, Torpdahl M, Nielsen EM, Vardund T, Aas L, Kapperud G. Harmonization of the multiple-locus variable-number tandem repeat analysis method between Denmark and Norway for typing Salmonella Typhimurium isolates and closer examination of the VNTR loci. J Appl Microbiol 2007; 102(3): 728–35.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[6] Torpdahl M, Sorensen G, Lindstedt BA, Nielsen EM. Tandem repeat analysis for surveillance of human Salmonella Typhimurium infections. Emerg Infect Dis 2007; 13(3): 388–95.
| PubMed |



