212 HETEROGENEITY OF RIBOSOMAL RNA GENE ACTIVATION AMONG CELLS OF IN VITRO-PRODUCED PORCINE EMBRYOS
B. Bjerregaard A , F. Strejcek B C , Z. Rasmussen D , J. Laurincik B C , H. Niemann E , P. Maddox-Hyttel A and P.D. Thomsen AA Department of Animal and Veterinary Basic Sciences, Royal Veterinary and Agricultural University, 1870 Frederiksberg C, Denmark
B Constantin the Philosopher University, Nitra, Slovak Republic
C Research Institute of Animal Production, Nitra, Slovak Republic
D Department of Small Animal Clinical Science, 1870 Frederiksberg C, Denmark
E Department of Biotechnology, Institute for Animal Science (FAL), Mariensee, 31535 Neustadt, Germany. Email: bol@kvl.dk
Reproduction, Fertility and Development 17(2) 256-257 https://doi.org/10.1071/RDv17n2Ab212
Submitted: 1 August 2004 Accepted: 1 October 2004 Published: 1 January 2005
Abstract
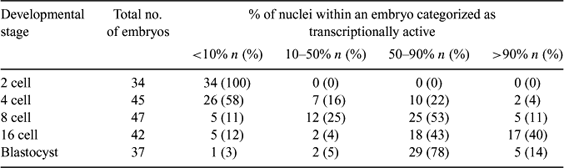
In vitro production (IVP) of porcine embryos by in vitro maturation of oocytes followed by fertilization and culture in vitro is hampered by great deficiencies. Initiation of at least the major embryonic genome transcription, which includes activation of ribosomal RNA (rRNA) genes and the associated formation of a fibrillo-granular nuclealus, is normally seen during the 4-cell stage in pigs. We have investigated the activation of rRNA synthesis and the presence of silver staining nucleolar proteins in porcine IVP embryos as a marker of transcriptional activity and, thus, developmental competence. A total of 205 porcine IVP embryos from the 2-cell to the blastocyst stage were examined using sequential fluorescent in situ hybridization (FISH) to the rRNA genes and their transcripts and silver staining of nucleolar proteins as previously described (Viuff et al. 2002 Biol. Reprod. 66, 629–634). Briefly, cumulus-oocyte complexes with at least three cumulus cell layers and evenly granulated ooplasm were isolated from 2–5 mm ovarian follicles with stereomicroscopic evaluation. Subsequently, oocytes were matured in NCSU-37 and mechanically denuded followed by fertilization using frozen-thawed epididymal semen. Presumptive zygotes were then cultured in NCSU-23 at 39°C, 5% CO2. Around the time of expected cleavage, the embryos were examined every second hour to determine the time of cleavage. Embryos at the 2-cell stage were harvested at 5 h post-cleavage (hpc), 4-cell embryos late during the third cell cycle at 30 hpc, and tentative 8- and 16-cell embryos at 10 hpc. Blastocysts were harvested at Day 5 post-insemination. In general, nuclei of 2-cell embryos displayed 4 small foci of FITC labelling (presumably the rDNA), but no specific silver staining, and were consequently categorized as transcriptionally inactive. At the late 4-cell stage, 58% of the embryos resembled the 2-cell stage. However, in the remaining embryos (42%), some or all nuclei displayed large areas of FISH labelling (presumptive rDNA and rRNA) co-localized with silver staining, and were catagorized as transcriptionally active. Among the 8-cell embryos, 64% displayed a majority of transcriptionally active nuclei, whereas this was the case in 83% and 92% of the embryos in the 16-cell embryos and the blastocysts, respectively. In general, the majority of the embryos contained a mixture of transcriptionally active and inactive cells. These findings show that the porcine IVP embryos are often delayed and asynchronous with respect to activation of the rRNA genes.
|
This work was supported by grants from “Disease models, disease prevention and animal welfare improvement: The pig embryo as a model.” Danish Research Agency (Grant: 9901178), NATO (Grant: 978658), and Deutsche Forschungsgemeinschaft (DFG).


