264 IDENTIFICATION OF THE PREDOMINANT TRANSCRIPT IN BOVINE OOCYTES, IN VITRO-DERIVED BLASTOCYSTS, AND IN NUCLEAR TRANSFER BLASTOCYSTS
R. S. Prather A , S. Korte A , R. Woods A , L. Spate A , N. Bivens A , L. J. Forrester A , G. K. Springer A , R. V. Patel A , Z. H. Liu A , C. N. Murphy A and J. A. Green ADepartment of Animal Sciences, Univeristy of Missouri-Columbia, Columbia, MO 15201, USA
Reproduction, Fertility and Development 18(2) 239-240 https://doi.org/10.1071/RDv18n2Ab264
Published: 14 December 2005
Abstract
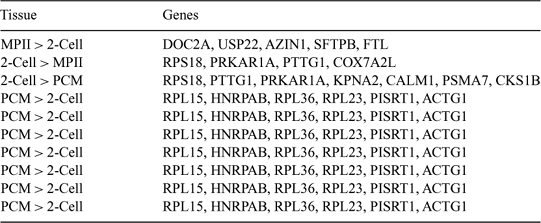
Identification of transcripts produced during bovine embryogenesis is the first step in describing the normal developmental program. To that end, mRNA was isolated from in vitro-matured metaphase II oocytes (MPII), in vitro-produced 2-cell-stage (2-Cell), in vitro-produced precompact morula-stage (PCM), in vitro-produced blastocyst-stage (BL), and in vitro-produced nuclear transfer blastocyst-stage (NTBL) embryos. The mRNA was isolated by using Dynabeads® (Dynal, Inc., Lake Success, NY, USA), and amplified by using the SMART system. PCR products were purified and ligated into pSPORT1 and electroporated into E. coli. Random clones were selected for DNA sequencing. Sequence data were evaluated for quality and clustered by sequence similarity with sequences generated from a larger expressed sequence tag (EST) project (http://genome.rnet.missouri.edu/Bovine/) by using the tlcluster program from the University of Iowa. Sequences over 100 bp in length with average Phred scores of over 20 for the entire sequence were submitted to GenBank (NIH genetic sequence database). Sequences were compared to the bovine TIGR (The Institute for Genomic Research) and human databases to gather annotation. The best comparison is listed below by using the HUGO Gene Nomenclature Committee standards (http://www.gene.ucl.ac.uk/nomenclature/) when possible. The number of unique clusters, i.e. no match in GenBank, was 53, 120, 109, 115, and 135, for MPII, 2-Cell, PCM, BL, and NTBL, respectively. The total number of clusters per tissue ranged from 224 to 992. The percent of clusters (number of clusters per total number of ESTs) per library was 12% (224/1762), 42% (746/1771), 48% (819/1715), 49% (900/1818) and 53% (992/1876) for MPII, 2-Cell, PCM, BL, and NTBL, respectively. Either the quality of the MPII library was lower or the complexity of the MPII mRNA was less than mRNA in the other tissues. Examples of mRNA that were in different abundance are shown in Table 1. Clearly, as in other species, there are significant changes in mRNA abundance during early embryogenesis. Furthermore, NTBL embryos, even though they are morphologically similar to BL, possess a population of mRNA that is distinct from that in BL.
|
This work was funded by the USDA NRI 2003–35205–12812 and Food for the 21st Century.


