228 microRNA REGULATION OF GENES IN BOVINE OOCYTES AND EMBRYOS
J. P. Barfield A , G. J. Bouma A and G. E. Seidel Jr AColorado State University, Fort Collins, CO, USA
Reproduction, Fertility and Development 22(1) 272-272 https://doi.org/10.1071/RDv22n1Ab228
Published: 8 December 2009
Abstract
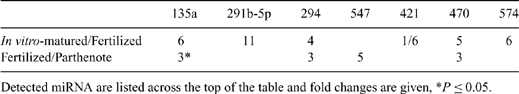
Little is known about expression of microRNA (miRNA) in bovine oocytes and pre-implantation embryos. These molecules likely have an important role in regulating development. For example, differences in quality of oocytes matured in vivo v. in vitro might be due, in part, to altered miRNA expression. In Experiment 1, in vivo-matured COC were collected by transvaginal aspiration of 7 superstimulated cows 21 to 23 h after GnRH injection, given 48 h after prostaglandin F2α and the last of 6 FSH injections given b.i.d. Oocytes aspirated from abattoir ovaries were matured in vitro for 23 h in a chemically defined medium. After vortexing, maturation of both groups of oocytes was confirmed by visualization of the first polar body, and oocytes were snap frozen in mirVana lysis buffer (Applied Biosciences, Foster City, CA, USA). In Experiment 2, in vitro-matured oocytes were generated as described. Subsets were fertilized in vitro or activated parthenogenetically by incubation in 5-μM ionomycin for 5 min followed by 10 μg mL-1 cycloheximide plus 5 μg mL-1 cytochalasin B for 5 h. After 18 h and 12 h, respectively, fertilized and activated oocytes were centrifuged at 10 000 × g for 10 min to enable visualization of pronuclei. Zygotes with 2 polar bodies and 2 pronuclei and parthenotes with 2 pronuclei were snap frozen in mirVana lysis buffer. Total RNA was extracted from 30 pooled oocytes for each replicate using the mirVana MiRNA Isolation Kit (Ambion, Inc., Austin, TX, USA). Reverse transcription of RNA was performed using the QuantiMir RT kit (System Biosciences, Mountain View, CA, USA), and miRNA expression was evaluated by real-time PCR using the Mouse miRNome Profiler plate, which contains primers for 384 miRNA (System Biosciences). Three plates were analyzed for each group (30 oocytes per plate). Changes in relative expression levels were analyzed with a t-test of values normalized to miR-181a, which was consistently expressed in all samples. In Experiment 1, compared with in vitro-matured oocytes, in vivo-matured oocytes had 11-fold higher (P = 0.02) expression of miR-375, which targets numerous genes involved in electron transport chain and oxidative phosphorylation pathways according to the bioinformatic database mirGator. MiR-291a-5p, miR-494, miR-539, and miR-547 were expressed in in vivo-matured oocytes only; the converse was found for miR-575-5p. Results from Experiment 2 are in the table. Major pathways associated with potential targets of the detected miRNA include TGF-beta signaling, Wnt signaling, tight junction formation, DNA replication reactome, steroid biosynthesis, mRNA processing binding reactome, and glutamate metabolism. Several of these candidate miRNA might be important for regulation of bovine oocyte maturation and embryo development.
|


