Phylogenomics of the green ash eucalypts (Myrtaceae): a tale of reticulate evolution and misidentification
Susan Rutherford A B C , Peter G. Wilson B , Maurizio Rossetto B and Stephen P. Bonser AA Evolution and Ecology Research Centre, School of Biological, Earth and Environmental Sciences, UNSW Australia, Sydney, NSW 2052, Australia.
B National Herbarium of NSW, Royal Botanic Garden Sydney, Mrs Macquaries Road, Sydney, NSW 2000, Australia.
C Corresponding author. Email: susan.rutherford@rbgsyd.nsw.gov.au
Australian Systematic Botany 28(6) 326-354 https://doi.org/10.1071/SB15038
Submitted: 3 September 2015 Accepted: 7 December 2015 Published: 10 May 2016
Abstract
Eucalyptus is a genus that occurs in a range of habitats in Australia, Papua New Guinea, Timor, Sulawesi and the Philippines, with several species being used as sources of timber and fibre. However, despite its ecological and commercial significance, understanding its evolutionary history remains a challenge. The focus of the present study is the green ashes (subgenus Eucalyptus section Eucalyptus). Although previous studies, based primarily on morphology, suggest that the green ashes form a monophyletic group, there has been disagreement concerning the divergence of taxa. The present study aims to estimate the phylogeny of the green ashes and closely related eucalypts (37 taxa from over 50 locations in south-eastern Australia), using genome-wide analyses based on Diversity Arrays Technology (DArT). Results of analyses were similar in topology and consistent with previous phylogenies based on sequence data. Many of the relationships supported those proposed by earlier workers. However, other relationships, particularly of taxa within the Sydney region and Blue Mountains, were not consistent with previous classifications. These findings raise important questions concerning how we define species and discern relationships in Eucalyptus and may have implications for other plant species, particularly those with a complex evolutionary history where hybridisation and recombination have occurred.
Additional keywords: Australia, DArT, Diversity Arrays Technology, Eucalyptus, hybridisation, phylogenetics, recombination.
Introduction
Eucalyptus L’Hér. (Myrtaceae) is a highly diverse genus encompassing more than 700 species distributed across Australia, Papua New Guinea, Timor, Sulawesi and the Philippines (Smith et al. 2003; McKinnon et al. 2008; Wilson 2011). Over 98% of species within the genus are endemic to Australia where they are the dominant or co-dominant component of many vegetation types (Potts and Wiltshire 1997; Hager and Benson 2010). Eucalyptus is also commercially important, with many species (such as Eucalyptus grandis W.Hill, E. globulus Labill. and E. tereticornis Sm.) being grown around the world as sources of timber and fibre (Eldridge et al. 1993; Grattapaglia et al. 2012). Eucalypts are considered Gondwanan in origin (Crisp et al. 2011; Gandolfo et al. 2011; Hermsen et al. 2012; Thornhill and Macphail 2012), forming a minor part of Tertiary Australian rainforests (Hill 1994; Hager and Benson 2010). Macrofossil evidence suggests that the distribution of eucalypts expanded in response to increasing aridity during the Miocene, and pollen evidence indicates that they became widespread only in the Pleistocene (5–1.5 million years ago; Pole et al. 1993; Rozefelds 1996; Potts and Pederick 2000; Macphail 2007). Many present-day eucalypt species complexes are thought to be the result of recent and ongoing speciation (McKinnon et al. 2004; Byrne 2007; Yeoh et al. 2013). Morphological differences among species are often narrowly defined (Hill 1991), and clinal variation and morphological convergence between taxa are common (McKinnon et al. 2004). Defining species boundaries is further complicated by interspecific hybridisation, often between distantly related taxa (Griffin et al. 1988; Rossetto et al. 1997; McKinnon et al. 2001; Field et al. 2011a, 2011b; Steane et al. 2011; Pollock et al. 2013, 2015). As a result, understanding evolutionary relationships in Eucalyptus, particularly between closely related species, remains a major challenge.
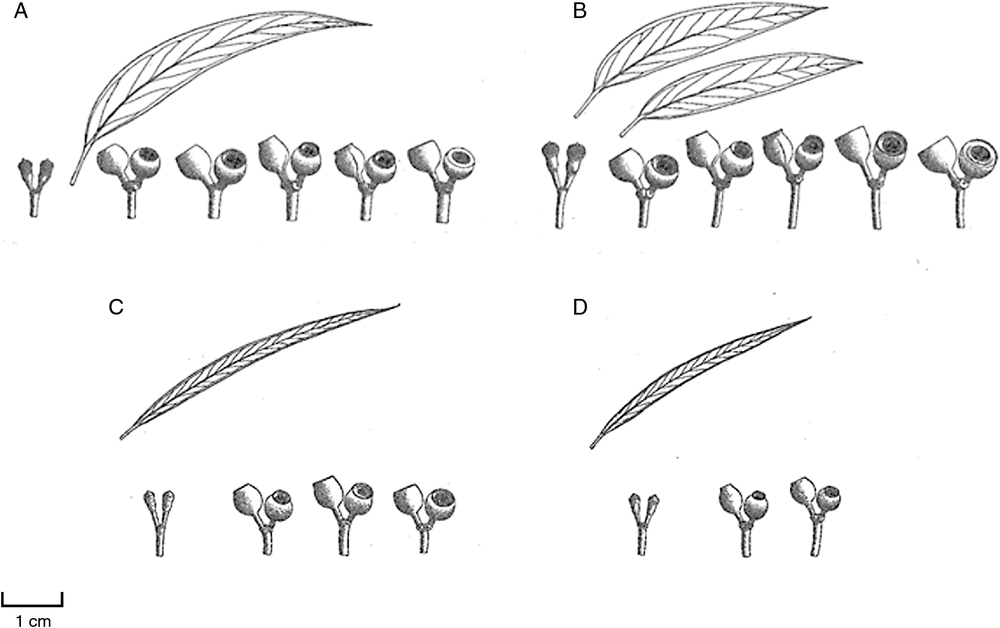
The focus of the present study is the green ashes in subgenus Eucalyptus section EucalyptusA (Brooker 2000). The green ashes are characterised by alternate juvenile leaves, adult leaves with moderate to no reticulation, pedicellate buds, reniform anthers and brown to red–brown seeds (Brooker 2000). They are found in a range of habitats in south-eastern Australia, with some species occurring as trees in tall forests on fertile soils and others as small trees or mallees on shallow soils on sandstone (Ladiges et al. 2010). Thirteen species were recognised by Brooker (2000), including Eucalyptus regnans (the tallest flowering plant in the world, up to 100 m tall), the timber species, E. obliqua and E. fastigata, and the mallee, E. cunninghamii, which is often less than 1 m in height (Fig. 1). Of these, nine are rare, restricted or localised (e.g. E. paliformis is known from only seven populations in Wadbilliga National Park, Prober et al. 1990). Previous studies, based primarily on morphology, suggest that the green ashes form a monophyletic group (Ladiges et al. 1987, 1989). However, there has been much disagreement concerning the divergence and differentiation of taxa, and, in particular, the number of recognised species (Table 1). A small number of species, namely E. regnans, E. fastigata E. obliqua, E. triflora, E. obtusiflora, E. stricta, E. apiculata, E. kybeanensis and E. approximans, were placed as part of series Obliquae (section Renantheria) by Pryor and Johnson (1971). The green ashes were only later referred to as a ‘group’ (e.g. Ladiges et al. 1987, 1989; Moran et al. 1990; Prober et al. 1990; Hill 1991, 2002). Ladiges et al. (1989) recognised more species (e.g. E. dendromorpha, E. rupicola, E. paliformis and E. burgessiana), although they considered E. obtusiflora (now E. obstans) to be a subspecies of E. stricta. Hill (1991, 2002) treated several taxa as species, e.g. E. codonocarpa (formerly E. approximans subsp. codonocarpa), E. spectatrix, E. laophila and E. microcodon, and recognised E. langleyi, E. obstans (formerly E. obtusiflora) and E. cunninghamii (formerly E. rupicola). However, many of the species recognised by Hill (1991, 2002) were not recognised by Brooker (2000) (Fig. 2). In the green ashes, Brooker and Kleinig (2006) and Slee et al. (2006) considered E. obstans to be a coastal variant of E. burgessiana, E. spectatrix to be a southern outlier of E. stricta, E. laophila to be a synonym of E. apiculata, and include both E. codonocarpa and E. microcodon within E. approximans subsp. codonocarpa. Many of these species are very similar morphologically and are difficult to distinguish in the field (Lassak and Southwell 1982; Ladiges et al. 1989). Consequently, although much effort has gone into the systematics and classification of this group, the ranking of taxa and the nature of the relationships among species remain uncertain.

|
|
Over the past two decades, molecular methods have become increasingly important in resolving questions concerning evolutionary relationships among taxa. The development of sequence datasets has enhanced our understanding of relationships between eucalypt genera and major subgenera (e.g. Udovicic et al. 1995; Steane et al. 1999, 2002; Udovicic and Ladiges 2000; Whittock et al. 2003; Parra-O. et al. 2006, 2009; Ochieng et al. 2007b). However, although standard DNA markers have successfully been used to resolve relationships at higher taxonomic levels within Eucalyptus, these have generally been unsuccessful in resolving relationships among closely related species (McKinnon et al. 2008). Most phylogenetic studies use sequence data from only a single or few regions of the genome (e.g. chloroplast DNA) and are not sufficiently variable in closely related species (e.g. Steane et al. 1998). There are also problems associated with using sequence data from some regions of nuclear DNA (e.g. ITS and ETS) in eucalypts because of the functional constraints imposed on neutral change of nucleotides during evolution (Bayly and Ladiges 2007; Ochieng et al. 2007a; Bayly et al. 2008). Marker systems theoretically representing the whole genome, such as microsatellites and amplified fragment length polymorphisms (AFLPs), have been used to overcome some of these issues (Steane et al. 2011). However, although microsatellites have moderate levels of throughput, and are highly polymorphic and transferable across populations, their transferability across species is sometimes poor (Rossetto et al. 2000; Semagn et al. 2006). The genotyping density obtained even with AFLPs is only hundreds of markers per sample and because it is a gel-based technique, it is comparatively labour intensive (Sansaloni et al. 2010).
With the advent of next generation sequencing (NGS), analytical approaches that have wider genome coverage have been developed. Bayly et al. (2013) used whole chloroplast genome sequences to construct a phylogeny of 39 eucalypt species, with many branches having 97–100% bootstrap support. Another technique that has recently been used in Eucalyptus is Diversity Arrays Technology (DArT) (Hudson et al. 2012). DArT is a microarray hybridisation-based technique that simultaneously assays hundreds to thousands of markers across the genome (Jaccoud et al. 2001; Sansaloni et al. 2010; Kullan et al. 2012). Steane et al. (2011) used over 8000 DArT markers (primarily nuclear) to construct a phylogeny of Eucalyptus, where relationships among higher taxa were generally concordant with traditional taxonomy and ITS-based phylogenies, with high resolution within major clades (including between some closely related species) relative to previous techniques.
Although several green ash taxa (e.g. Eucalyptus regnans, E. obliqua, E. triflora) have been included in molecular phylogenies over the past 10 years (Bayly and Ladiges 2007; Steane et al. 2011; Bayly et al. 2013), there has been no broader study of the green ash eucalypts using these more advanced techniques. Prober et al. (1990) used allozyme data to investigate diversity in the green ashes, and this revealed low differentiation among taxa and many relationships that were not consistent with those derived from morphological characters. Although the green ashes are widely distributed in south-eastern Australia, they are particularly diverse in the Sydney region and Blue Mountains (the latter was listed as a World Heritage Area partly because of its eucalypt diversity (Hager and Benson 2010). Within this area, the green ashes are distributed across a range of environments and occur sympatrically with other closely related eucalypts, such as blue ashes (including scribbly gums), black sallies, stringybarks and peppermints (sections Cineraceae, Longitudinales, Capillulus and Aromatica respectively; Brooker 2000). The distribution of taxa in this heterogeneous environment, therefore, provides a unique opportunity for using more recent genomic techniques to address specific evolutionary questions concerning the green ashes and closely related taxa. Our objective was to estimate the phylogeny of the green ashes using DArT markers, so as to resolve relationships within the green ash group and between the green ashes and other taxa in subgenus Eucalyptus. Therefore, we aimed to address the following questions: (1) do the green ashes form a monophyletic group, (2) is there evidence of hybridisation among taxa, (3) are phylogenetic relationships of the green ashes and closely related taxa consistent with previous classifications (primarily based on morphological characters), and (4) are phylogenetic relationships correlated with geography and substrate?
Materials and methods
Sampling of taxa
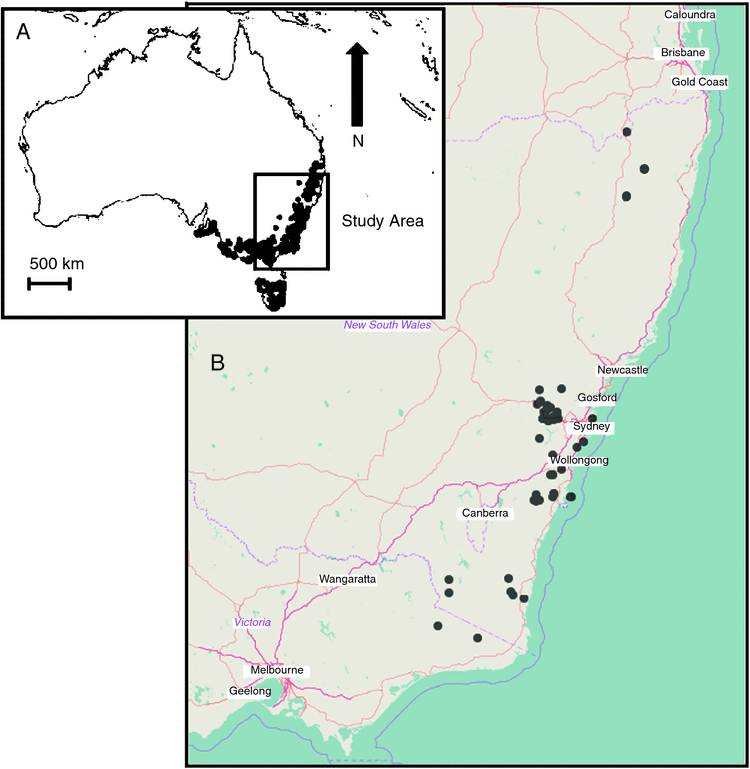
Leaf material was collected from all taxa assigned to the green ash group by the major authorities (Pryor and Johnson 1971; Ladiges et al. 1989; Hill 2002; Brooker 2000). Table 2 lists the species sampled, following the species concepts of Hill (2002). For most of these species, more than one individual was sampled from multiple locations. So as to sample across the diversity and geographic range of the group, we collected from 44 locations between southern Queensland and Victoria (Fig. 3). Locations of green ash taxa and habitat details were obtained from the National Herbarium of New South Wales database (Royal Botanic Garden Sydney) and Benson and McDougall (1998) (full accession details are listed in Appendix 1; habitat details, and latitude and longitudes are provided in Appendix 2). During the sampling, several new populations of green ash taxa (e.g. Eucalyptus stricta) were discovered and included. In addition, closely related co-occurring taxa in subgenus Eucalyptus (sections Aromatica, Capillulus, Cineraceae and Longitudinales) were sampled (often from more than one individual per species from different locations). Eucalyptus cloeziana (subgenus Idiogenes) was included as an out-group to subgenus Eucalyptus on the basis of previous studies (Sale et al. 1993; Hill and Johnson 1995; Ladiges et al. 1995; Steane et al. 1999; Udovicic and Ladiges 2000; Steane et al. 2011). Most taxa were sampled directly in the field and their geographic position (including elevation) was recorded (GPS model: Garmin Rino 650, Garmin Australasia, Sydney, NSW, Australia); vouchers of these were lodged in the National Herbarium of New South Wales. Other species (namely E. approximans, E. regnans, E. deuaensis, E. caliginosa, E. cloeziana and E. apiculata from the Berrima population) were sourced from specimens cultivated at the Currency Creek Arboretum (South Australia), the Royal Botanic Garden Sydney, the Australian Botanic Garden (Mount Annan) and the Blue Mountain Botanic Garden (Mount Tomah). All leaf samples were dried in silica gel and stored at −20°C until used for DNA extraction.

|
|
DNA isolation
Total genomic DNA was extracted from samples using a CTAB protocol modified from Doyle and Doyle (1990). A total of 1–1.5 g of leaf material per sample was ground under liquid nitrogen and the following modifications were made: (1) 2-mercaptoethanol was replaced by sodium metabisulfite (0.5%); (2) addition of sorbitol (0.35 M), polyvinylpyrollidone (4%) and sarcosyl (5%) to the CTAB isolation buffer; and (3) DNA was purified using a Zymo-Spin I-96 Plate and the ZR-96 Clean and Concentrator Kit (Zymo Research Corporation, CA, USA). DNA quality of each sample was tested by restriction of 2 µL of DNA with 3 µL of the restriction endonuclease, RsaI (New England Biolabs, Irvine, CA, USA), and digests were visualised on a 1.0% agarose gel. DNA concentrations were measured using a Qubit 2.0 Flourometer (Invitrogen, Melbourne, Vic., Australia) and each sample was made up to between 400 and 1000 ng of DNA (targeting a concentration of 50 ng µL–1). Samples were sent to Diversity Arrays Technology Pty Ltd (Canberra, ACT, Australia) for genotyping, using the microarray platform developed by Sansaloni et al. (2010).
Phylogenetic analysis of DArT markers
The DArT microarray genotyping platform produces a binary output showing the marker name, its presence or absence in each sample and statistics regarding the quality and reliability of each marker. The DArT dataset produced for the present study consisted of a total of 2702 presence or absence markers. Phylogenetic trees were constructed using parsimony, Bayesian and distance analyses. To ensure that only the higher-quality markers were used, markers with a call rate below 90% and reproducibility less than 100% were removed from the dataset (leaving 1780 markers). Maximum parsimony (MP) analyses were conducted in PAUP 4.0 b10 (D. L. Swofford, Sinauer, Sunderland, MA, USA). The MP analysis was performed with a heuristic search using 1000 random addition sequences and tree bisection and reconnection (TBR) branch swapping (characters were equally weighted, gaps were treated as missing and character states were unordered). Bootstrapping for the MP analysis (branch lengths shown) comprised heuristic searches and 1000 replicates. Bayesian analyses were conducted in MrBayes 3.2.4 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2012) using a restriction site (binary) model of evolution and default priors. The final analysis was run for 150 million generations sampling every 1000 generations, with two parallel runs each with four chains (three hot and one cold). Convergence was considered reached on the basis of the standard deviation of split frequencies (<0.01) and the first 25% of trees were discarded as burn-in.
Diversity Arrays Technology (DArT) datasets are considered to follow a Dollo model of evolution (because it is much easier for a DArT marker to be lost than gained; Woodhams et al. 2013). Although Dollo data have been traditionally analysed using parsimony methods (Le Quesne 1974; Farris 1977), it is well known that parsimony does not take into account branch-length information (Woodhams et al. 2013). Therefore, a distance-based phylogenetic approach, which implements the Dollo model of evolution, was used in the present study. A distance matrix of the DArT data (Partitioned Additive Dollo Distance, or PADD) was calculated following the method outlined by Woodhams et al. (2013) and a tree found by minimum evolution in FastME (Desper and Gascuel 2002; Lefort et al. 2015). Branch support was obtained using a bootstrap analysis in PAUP 4.0 b10 (D. L. Swofford). This comprised a heuristic search and 1000 replicates (under the minimum-evolution criterion). Nexus files containing the raw data and all tree files are available on TreeBase at http://purl.org/phylo/treebase/phylows/study/TB2:S18461 (accessed 9 November 2015).
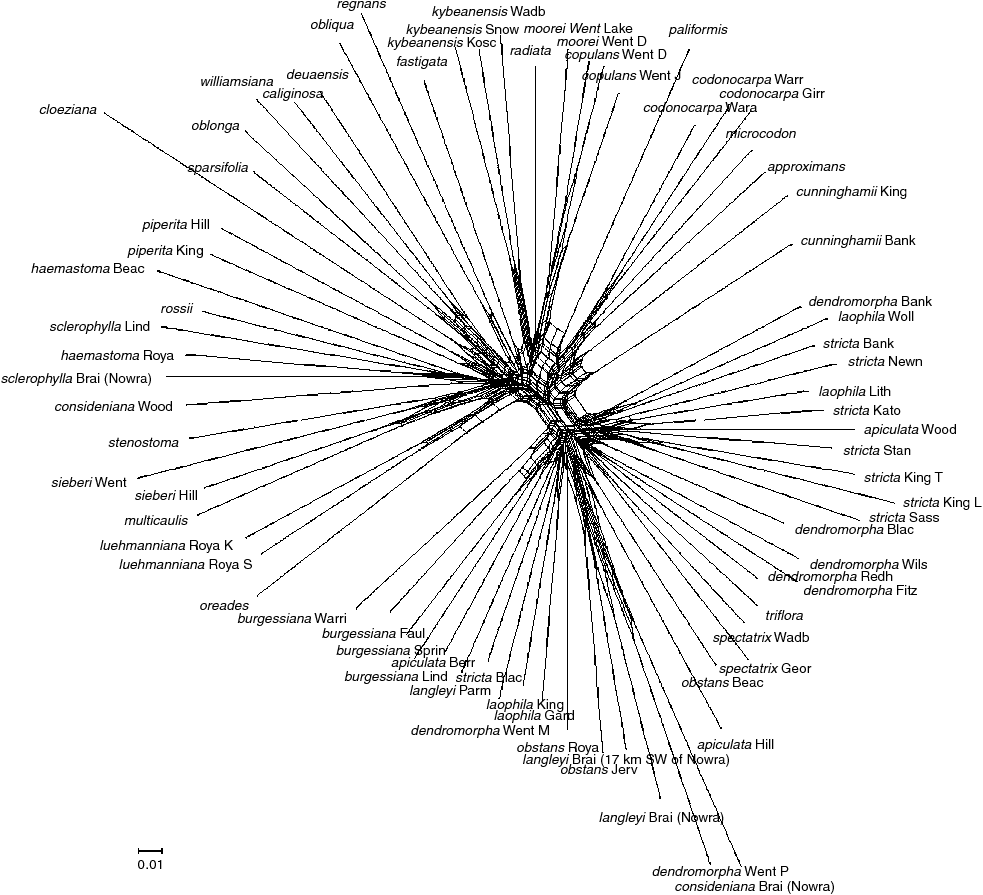
Relationship networks based on the full DArT dataset (2702 markers) were generated in SplitsTree4 (version 4.13.1) (Huson 1998; Huson and Bryant 2006) using the default settings of the software. Relationship networks are implicit representations of evolutionary history that are used to represent agreement and incompatibilities in the dataset (Huson and Bryant 2006). Therefore, use of the full DArT dataset for these analyses was considered appropriate. In a relationship network, the parallel edges indicate splits in the data and allow samples to be assigned to groups, with the longer lines suggesting more support for that particular split (Huson and Bryant 2006). Relationship networks are an effective way of depicting the character conflicts of DArT markers and allow the complexity of the datasets to be visualised (Steane et al. 2011).
Reconstruction of ancestral states and character evolution
To examine patterns and variation in morphology, ancestral reconstructions were performed on the following diagnostic traits: (1) habit (mallee or tree) and (2) leaf width. These parameters were chosen as they are considered important when identifying species in subgenus Eucalyptus in the classifications of Brooker (2000) and Hill (2002). Leaf width was measured at the widest point (following the method of McGowen et al. 2001) from five random leaves per voucher specimen to the nearest millimetre, with a digital Vernier calliper (Kincrome, Melbourne, Victoria, Australia). For Eucalyptus microcodon and E. williamsiana, vouchers from the same population were used for leaf-width measurements (because of the unavailability of leaves from the samples used for DNA analysis). Categories for leaf length and leaf width have not been standardised in eucalypts. However, in the treatment of Hill (2002), E. stricta is described as narrow-leaved (with leaves <10 mm wide), whereas E. burgessiana is described as broad-leaved (leaf width >15 mm). Therefore, the categories used here for leaf width were based on the descriptions of Hill (2002) and divided into narrow (<10 mm), intermediate (10–15 mm) and broad (>15 mm). The contribution of two environmental variables (altitudinal zone and substrate) to the evolutionary diversification of taxa was also investigated. Altitudinal divisions followed the zones defined by Turak et al. (2011) and were classified as follows: coastal and lowland (0–235 m), upland (235–1065 m) and highland (≥1065 m). The substrate observed (sandstone, granite, basalt or rhyolite) was recorded per sample at each site at the time of collection of leaf material for DNA analysis. Ancestral reconstructions of each morphological and environmental parameter were traced onto the Bayesian phylogenetic tree by using MP reconstructions in the Mesquite software package v. 3.03 (W. P. Maddison and D. R. Maddison, see http://mesquiteproject.org). The character data matrix is presented in Appendix 2.
Results
Phylogenetic analysis of DArT markers
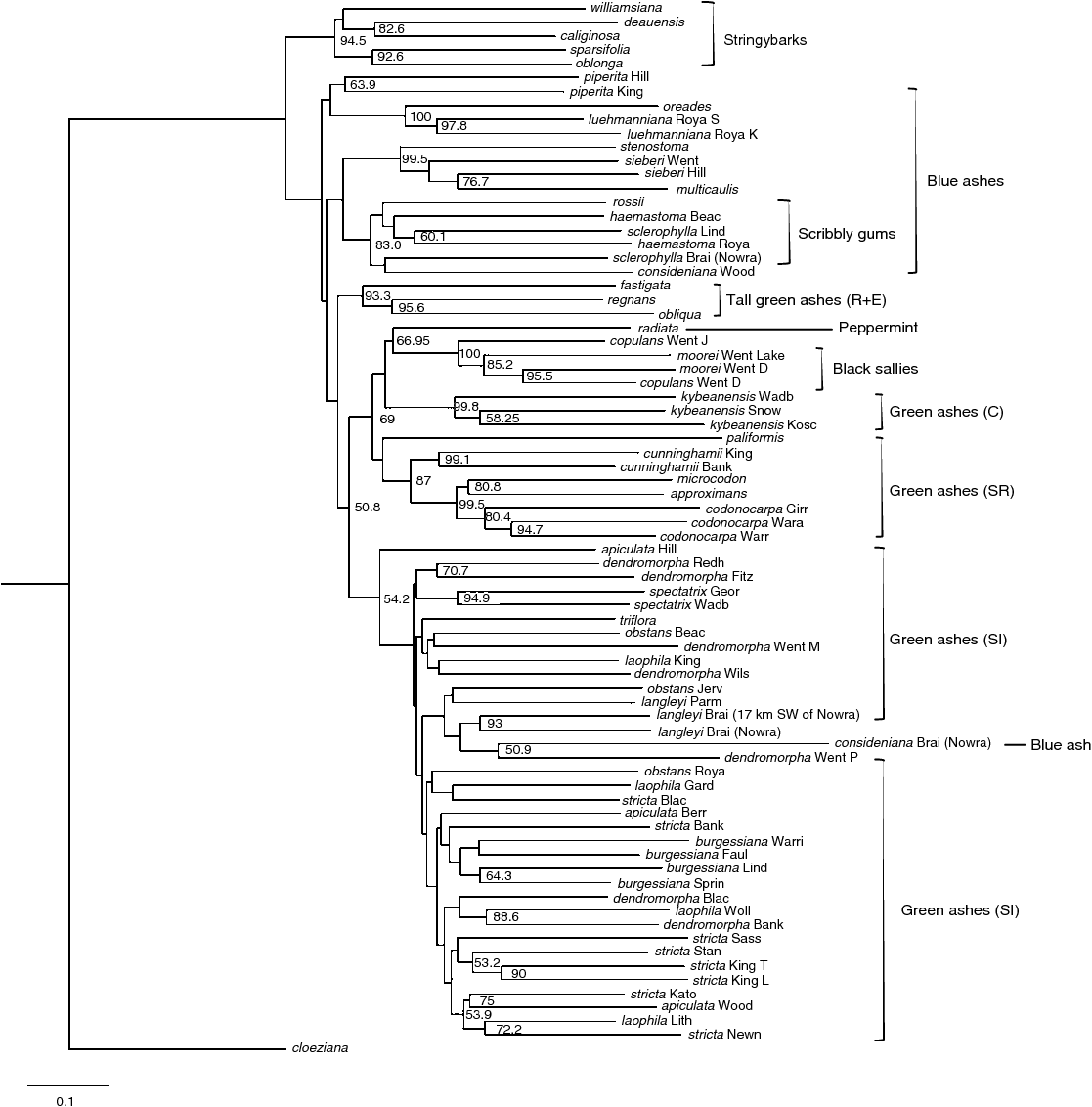
The DArT dataset used to produce phylogenetic trees (comprising 1780 markers) consisted of 76 samples (representing 37 taxa), with the proportion of missing data for most samples being less than 5%. Five samples had 5–10% missing data, whereas Eucalyptus regnans had the highest proportion of missing data (22%). The overall topology and groupings of taxa produced from all analyses were similar. The MP analysis recovered two trees, each with a tree length of 15 909, consistency index (CI) of 0.11, and retention index (RI) of 0.34. Of the 1780 markers in the dataset, 1695 were parsimony informative. The strict consensus tree had 74 nodes, 28 of which had bootstrap support (BS) greater than 50% (Fig. 4). Eucalyptus piperita (section Cineraceae) from two locations (Hilltop and Kings Tableland) was sister to the remainder of the taxa in subgenus Eucalyptus (although E. piperita did not form a monophyletic group). The stringybarks (section Capillulus) formed a monophyletic group (84.9% BS), which was sister to a clade comprising the green ash tall trees, E. regnans, E. obliqua and E. fastigata. Eucalyptus regnans and E. obliqua formed a clade (81.7% BS). The clade comprising the stringybarks and the green ash tall trees was sister to the remainder of the blue ashes (section Cineraceae), black sallies (section Longitudinales), the peppermint (section Aromatica) and the majority of the green ashes. The remainder of the blue ashes (apart from E. consideniana from Nowra) formed a monophyletic group comprising three main clades. The first was of E. oreades and E. luehmanniana (100% BS), the second included E. multicaulis, E. sieberi and E. stenostoma (99.3% BS), and the third comprised E. consideniana from Woodford and the scribbly gums (E. haemastoma, E. sclerophylla and E. rossii). Whereas the samples of E. luehmanniana emerged in a monophyletic group (76.9% BS), the samples of other species (namely E. sieberi, E. haemastoma and E. sclerophylla) did not form a clade.
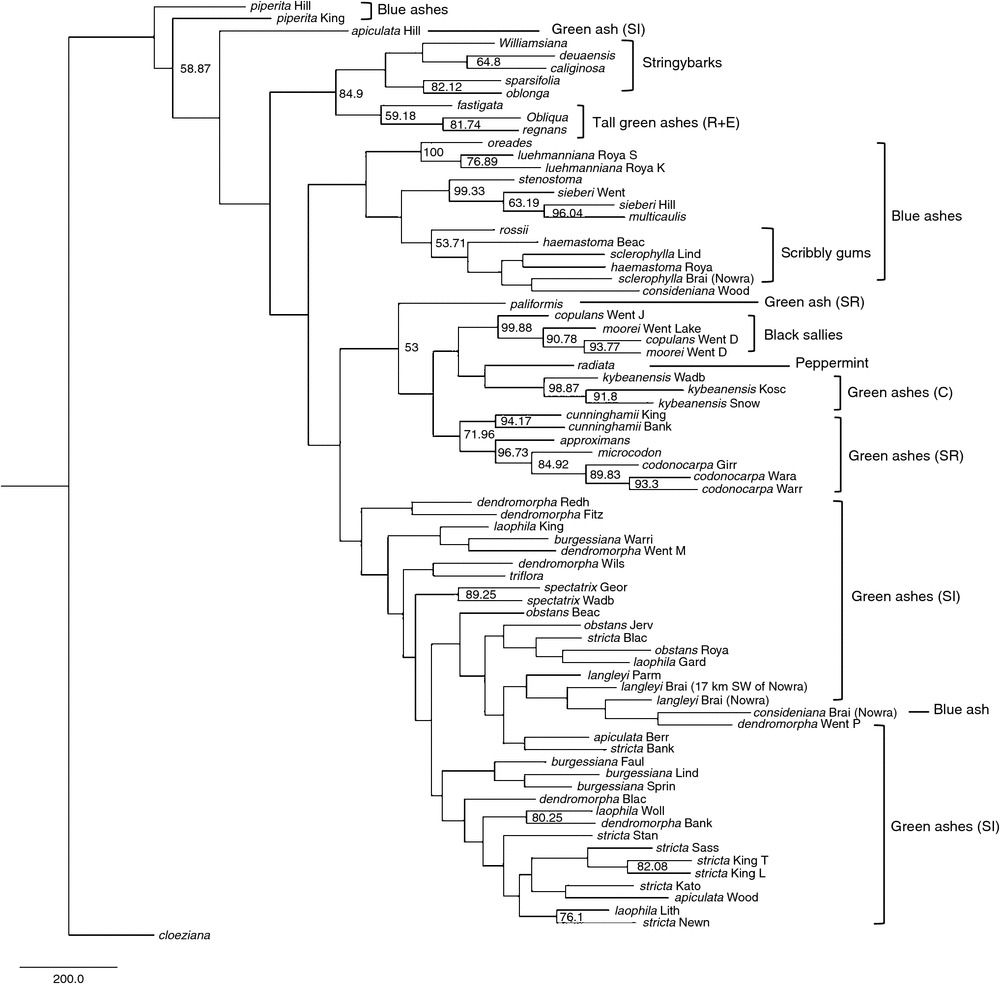
|
With the exception of Eucalyptus apiculata from Hilltop, the remainder of the green ashes, the black sallies, E. radiata and E. consideniana from Nowra formed a clade. Within this group were two main clades. The first comprised E. codonocarpa, E. approximans, E. microcodon, E. cunninghamii, E. kybeanensis, E. paliformis, E. radiata and the black sallies. Eucalyptus codonocarpa from all locations was monophyletic (89.8% BS), as were E. cunninghamii (94.2% BS) and E. kybeanensis (98.9% BS). The black sallies formed a monophyletic clade (99.9% BS); however, within this group, E. copulans from all locations and E. moorei from all locations did not form separate monophyletic clades. The second main clade included E. spectatrix, E. consideniana from Nowra and most of the green ash taxa from the Sydney region and Greater Blue Mountains World Heritage Area (GBMWHA). Eucalyptus spectatrix from both locations was monophyletic (89.3% BS), as was E. dendromorpha from Fitzroy Falls and Redhills Road, and E. burgessiana from three locations in the GBMWHA (Linden, Springwood and Faulconbridge). The three E. langleyi samples, E. dendromorpha from the Princes Rock track (Wentworth Falls, GBMWHA) and E. consideniana formed a clade. With the exception of E. stricta from Blackheath and Mount Banks, all other E. stricta populations emerged in a clade that also included E. apiculata from Woodford, E. laophila from Lithgow and Wollemi National Park, and E. dendromorpha from Mount Banks. Eucalyptus apiculata from Hilltop was separate from the other green ash taxa (being sister to all other taxa with the exception of E. piperita).
Bayesian analyses produced a phylogeny with 70 nodes, 49 of which had Bayesian posterior probability (PP) greater than 0.95 (Fig. 5). As in the MP analysis, E. piperita from Hilltop was sister to the remainder of taxa in subgenus Eucalyptus, and the green ash tall trees (E. regnans, E. obliqua and E. fastigata) formed a clade separate from the other green ashes (PP: 0.99). In contrast to the MP analysis, the remainder of the blue ashes were not monophyletic. However, as with the MP analysis, E. luehmanniana and E. oreades formed a monophyletic group (PP: 1), as did samples of E. multicaulis, E. sieberi and E. stenostoma (PP: 1). The remainder of the green ash taxa formed a clade with the black sallies and E. radiata (section Aromatica). This clade was split into the same two main groups as in the MP analysis. However, in contrast to the MP analysis, E. apiculata from Hilltop was grouped with E. spectatrix from southern New South Wales and the majority of green ash taxa from the Sydney region and GBMWHA (PP: 1). Also, unlike the MP analysis, all the E. burgessiana samples formed a monophyletic group (PP: 0.97); the E. langleyi samples used formed a monophyletic group (PP: 0.99); and the blue ash from Nowra, E. consideniana, was not grouped with E. langleyi, but was in a clade with the rest of the blue ashes.
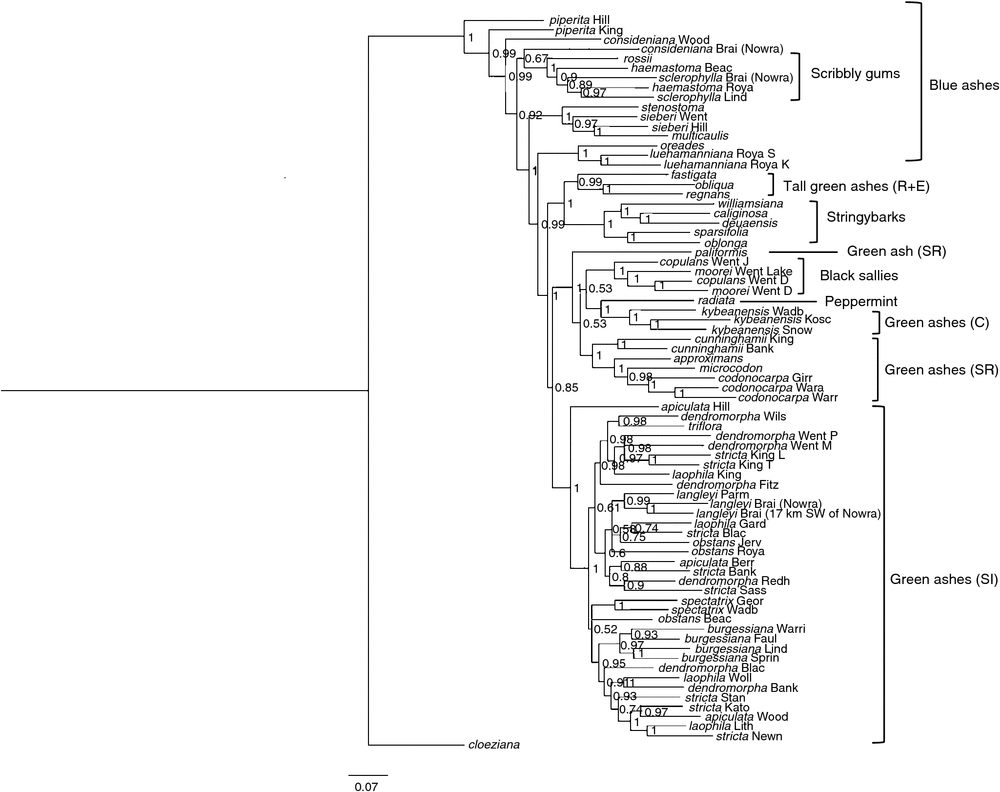
|
The minimum evolution tree produced in FastME from the PADD data had 74 nodes (38 of which had BS greater than 50%, Appendix 3). In contrast to the Bayesian and MP analyses, the two E. piperita samples were monophyletic (63.9% BS). Also, unlike the Bayesian and MP analyses, the stringybarks were sister to the remainder of taxa in subgenus Eucalyptus. However, the groupings of most taxa in this tree were similar to those in the Bayesian and MP trees. For example, as with the Bayesian and MP analyses, the green ash tall trees (E. regnans, E. fastigata and E. obliqua) were separate from the remainder of the green ashes (which formed a clade comprising the same two major groups). As in the Bayesian tree (but unlike the MP analysis), E. apiculata from Hilltop was sister to E. spectatrix and the green ashes from the Sydney region and GBMWHA. However, in contrast to the Bayesian tree (but like in the MP analysis), E. consideniana from Nowra was grouped with E. langleyi and E. dendromorpha from Princes Rock track (Wentworth Falls). Like in the Bayesian tree and unlike the MP tree, all samples of E. burgessiana were monophyletic.
Two relationship networks were generated using SplitsTree4, one including all taxa in subgenus Eucalyptus and the other comprising the green ash taxa only. In the relationship network comprising all samples, taxa formed the same broad groups as in the MP, Bayesian and PADD analyses (Appendix 4). The relationship network comprising the green ash taxa only (Fig. 6) was also largely in agreement with the MP, Bayesian and PADD analyses, and allowed geographic differentiation among taxa to be visualised (with the clustering of northern New South Wales and southern Queensland taxa and the clustering of southern New South Wales and northern Victorian taxa). The taxa from the Sydney region and GBMWHA generally clustered together, although Eucalyptus spectatrix from southern New South Wales was nested within this group (as indicated by all phylogenetic analyses). As with the Bayesian and PADD phylogenies, and in contrast to the MP analysis, E. apiculata from Hilltop was grouped with the other green ash taxa from the Sydney region and GBMWHA.

|
Reconstruction of ancestral states and character evolution
Overall, the Bayesian analysis produced a phylogeny that was most consistent with the current taxonomy of subgenus Eucalyptus (Brooker 2000). In the MP tree, E. apiculata from Hilltop was separate from the other green ashes and closer to the blue ash, E. piperita. Similarly in both the PADD and MP analyses, the blue ash, E. consideniana, from Nowra, was grouped close to the green ash, E. langleyi. Therefore, the Bayesian topology was selected for ancestral reconstructions.
The distribution of growth habit showed a marked dichotomy in the Bayesian 50% majority consensus phylogeny (Fig. 7). The deeper diverging clades of the phylogeny (and the ancestral habit of the green ashes) was reconstructed as the tree form. Eucalyptus luehmanniana and E. multicaulis represented the only change to mallee form in the blue ash group and E. deuaensis was the only change to mallee form in the stringybark group. Within the clades comprising the majority of mallees, there were very few reversions to the tree form. For example, E. triflora and E. dendromorpha (from Fitzroy Falls and Redhills Road) represented the only reversions to tree form in the clade comprising most of the Sydney and GBMWHA taxa. Patterns in leaf width were not as significant on the phylogeny as growth form, although narrow leaves appeared only in the green ash mallee group. Nevertheless, there were many reversions in this group to intermediate and broad leaves. There was some correlation between clades on the Bayesian 50% majority consensus tree and environmental parameters. The majority of taxa studied occurred in upland habitats on sandstone, with some clades radiating into lowland and coastal habitats on sandstone (e.g. E. langleyi and E. luehmanniana) and other clades radiating into upland or highland habitats on granite (taxa from northern New South Wales, southern Queensland, southern New South Wales and northern Victoria). The green ash tall trees (E. regnans, E. obliqua and E. fastigata) differed from all other groups being a clade on basalt, as did E. deuaensis, which was the only taxon to occur on rhyolite.

|
Discussion
Phylogenetic relationships and the monophyly of the green ashes
Bayesian, MP and PADD analyses produced phylogenies with similar topologies and groupings of taxa. The phylogenies produced here were more resolved than were previous phylogenies of subgenus Eucalyptus using traditional one-region sequence data (e.g. Steane et al. 1999, 2002; Bayly and Ladiges 2007). These findings demonstrate that phylogenetic analyses based on DArT markers can provide insights into evolutionary relationships among closely related species and groups that are taxonomically challenging. Although only a few taxa from the present study were included in more recent phylogenies, the relationships found here were generally consistent with the findings of Steane et al. (1999, 2002, 2011, for the relationship of Eucalyptus obliqua and E. regnans) and Bayly and Ladiges (2007, for the close relationship of E. triflora, E. spectatrix and E. paliformis). The results from the present study support many of the relationships proposed by Ladiges et al. (1989), Hill (2002) and Brooker (2000). The clade comprising E. williamsiana, E. deuaensis and E. caliginosa is consistent with Brooker’s (2000) section Capillulus and Hill’s (2002) stringybarks, whereas the close relationship of E. regnans and E. fastigata, and the relationship of the E. approximans–codonocarpa–microcodon clade with E. cunninghamii and E. paliformis is congruent with Ladiges et al. (1989) and Brooker (2000).
In the present study, the green ashes (subgenus Eucalyptus section Eucalyptus) as circumscribed by Brooker (2000) did not form a monophyletic group. The separation of the green ash tall trees from the remainder of the green ashes is in contrast to Brooker (2000) and Ladiges et al. (1989). The blue ashes (section Cineraceae) in the present study were polyphyletic, which disagrees with the classifications of Brooker (2000) and Hill (2002). Similarly, the positions of the black sallies (section Longitudinales) and E. radiata were unexpected. The monophyly of E. cunninghamii from both locations, E. kybeanensis from all locations and E. luehmanniana from both locations support the species circumscriptions of Brooker (2000). However, many taxa from different locations (e.g. E. haemastoma and E. sclerophylla and the majority of green ashes from the Sydney region and GBMWHA) do not appear to fit into the species delimitations of Ladiges et al. (1989), Hill (2002) or Brooker (2000). Consequently, these results highlighted the need for a potential revision of the infrageneric ranking of the green ashes, blue ashes, black sallies and peppermints. The implications of the findings from the present study to the taxonomy and classification of subgenus Eucalyptus are discussed in further detail below.
Gene flow and hybridisation
Some of the relationships found in the present study differed from a chloroplast genome study on eucalypts (Bayly et al. 2013), in which E. obliqua and E. radiata formed a monophyletic group, and E. sieberi and E. elata also formed a monophyletic group with E. regnans, all within the ‘Monocalypt’ clade (=subgenus Eucalyptus). Numerous studies have highlighted the issue of incongruence between phylogenies based on chloroplast and nuclear DNA (e.g. Soltis and Kuzoff 1995; Kim and Donoghue 2008; Wang et al. 2011; Yu et al. 2013; Govindarajulu et al. 2015) and, therefore, differences between the findings of Bayly et al. (2013) and the present study (based on DArT markers, which are predominantly nuclear) are not surprising. McKinnon et al. (1999) found extensive sharing of chloroplast DNA haplotypes among sympatric species from subgenus Eucalyptus in Tasmania, which showed a clear correlation with geographic patterns rather than phylogenetic relationships. Consequently, analyses using uni-parentally inherited markers alone may confound phylogenetic reconstruction in groups that frequently hybridise (McKinnon et al. 1999; Bayly and Ladiges 2007). However, McKinnon et al. (2010) found that, although E. globulus and E. cordata maintained strongly differentiated nuclear gene pools, leakage of nuclear DNA did occur between the two species (although cpDNA sharing was much more extensive).
In the present study, some of the relationships found were indicative of hybridisation and introgression between lineages. For example, in the PADD tree, the E. obstans sample from Jervis Bay was in a clade with E. langleyi (from which it is morphologically distinct but geographically proximate). In the MP and PADD analyses, one sample of the blue ash, E. consideniana, appeared in the same clade as E. langleyi from the same location (although it was grouped with the other blue ashes in the Bayesian phylogeny). Similarly, whereas the sample of E. apiculata from Hilltop was grouped with the other green ashes in the Bayesian and PADD trees, in the MP analysis it was separate from the remainder of the green ashes and closer to E. piperita. Comparisons made between this specimen and a specimen at the National Herbarium of New South Wales (Chippendale 1002, NSW327081) recorded as a likely hybrid between E. apiculata and E. piperita (from Berrima, which is geographically close to Hilltop) revealed similarities in leaf colour, shape and size. In addition, the fruit shape from the specimen used in the present study was more spherical, suggesting that it may be the result of hybridisation with E. piperita. Patterns of morphological variation and introgression of eucalypt species in Tasmania indicate that reticulate evolution occurred between divergent lineages during the Quaternary (McKinnon et al. 2004), and Hager and Benson (2010) suggested that such processes are likely to have played a major role in the evolutionary history of the green ashes of the GBMWHA. Future studies should, therefore, use both chloroplast and nuclear genomes to explore the role that reticulate evolution may have played in the evolution of this group. Ecological and phenological studies (e.g. differences in flowering time) focussing on sympatric populations and taxa may also provide insights into patterns of gene flow and hybridisation.
Classification, morphology and the issue of misidentification
Historically, species within subgenus Eucalyptus have been difficult to identify because many of the subgroups have few obvious distinguishing synapomorphic morphological characters (McKinnon et al. 1999). The green ashes exemplify this, with the majority of species being distinguished on the basis of characters such as leaf length, leaf width, fruit size and bud size (characters that can be variable across large geographic areas). In the present study, those taxa that have notably distinctive morphological traits or are geographically isolated tend to form well supported clades. For example, E. cunninghamii is easily identified on the basis of its small, soft-textured, silvery-green leaves, E. kybeanensis is distinguished on the basis of its conical or hemispherical fruits and sessile buds, whereas the E. approximans clade (including E. codonocarpa, and E. microcodon) is geographically disjunct.
However, with the exception of E. cunninghamii, the morphological traits used for species identification in the Sydney region and GBMWHA (such as leaf length and width) often overlap between taxa (the ancestral reconstructions of leaf width in the present study highlighted such overlaps between taxa). Furthermore, previous studies have demonstrated that such morphological traits can be highly plastic. For example, in Nothofagus cunninghamii, it was found that although leaf length and width partially depended on genotype, there was a significant effect of environmental factors on morphology (leaves became smaller and thicker with increasing altitude, Hovenden and Vander Schoor 2004). In the case of the green ashes, many taxa that are difficult to identify on the basis of morphology alone can be assigned to a particular taxon on the basis of geographical location. For example, E. laophila and E. apiculata from the GBMWHA are often distinguished on the basis of the elevation at which they occur (E. laophila is considered to occur at higher altitudes than is E. apiculata). Species definitions that are in large part based on geographical location have likely led to misidentifications, which is an obvious issue in phylogenetic reconstructions. Another problem with such taxa (which are synonymous in Brooker’s (2000) classification) is that there is the possibility that they are the one highly plastic species that has been distinguished on the basis of morphological differences that are not useful in species delimitations. However, although some taxa from the present study (e.g. E. copulans and E. moorei) would become a monophyletic clade if re-labelled according to the classification of Brooker (2000), re-labelling samples used for other taxa (e.g. E. apiculata and E. laophila) does not make them monophyletic in the MP, PADD or Bayesian phylogenies. Many studies have highlighted the importance of comparing physiology and anatomy with phylogenetic information to better understand evolutionary diversification in both plants and animals (e.g. Ackerly et al. 2000; Garland et al. 2005; Hodson et al. 2005). The relationship between genetic variation and physiological and anatomical traits of seedlings, juvenile and adult plants may, therefore, provide insights into the evolution of green ash taxa in the Sydney region and GBMWHA and should be the focus of future studies.
Geography, substrate and evolutionary models
Although the majority of taxa in the present study occupy upland habitats on sandstone, the ancestral reconstructions support the hypothesis of radiation of the green ashes and other taxa in subgenus Eucalyptus into a multitude of habitats, such as lowland and coastal habitats on sandstone, upland and highland habitats on granite, and upland habitats on basalt. There was also a correlation between habit and substrate (e.g. the tall green ash trees, Eucalyptus regnans, E. fastigata and E. obliqua, were found on basalt, whereas the smaller trees and mallees were found on sandstone or granite). The relationship networks in the present study indicated geographic structuring of many taxa and indicated that there is likely to be recombination, hybridisation and introgression. Previous studies have discussed the possibility that evolution in many eucalypts may not necessarily have been divergent (Chappill and Ladiges 1996; McKinnon et al. 2008) and that speciation in both plants and animals can occur during partial reproductive isolation (Wu 2001; Lexer and Widmer 2008; Mallet 2005). Although more traditional evolutionary models assume a tree, it is well known that more complex evolutionary scenarios (such as rapid radiation and reticulate evolution) are poorly described by these models (Huson and Bryant 2006; Morrison 2014). Phylogenetic networks, which allow horizontal reticulation events as well as vertical processes to be visualised, are increasingly being recognised as providing a more comprehensive picture of evolutionary history (Francis and Steel 2015). In the present study, the relationship networks suggested a complex pattern of evolution in the green ashes and closely related eucalypts. The role of environmental parameters (especially substrate and soil type) in the evolutionary diversification of these groups should be investigated. A detailed population-genomic study targeting taxa in the Sydney region and GBMWHA will also be required to better understand the complexity of evolution in the green ashes and to clarify species boundaries.
Consequences for the classification of Eucalyptus subgenus Eucalyptus
The classification of Brooker (2000) and draft scheme of Nicolle (2015) are largely in agreement with regard to the groupings of species considered in the present study (see Appendix 5 for a direct comparison between the two classifications). The major difference is in the ranking; Brooker (2000) recognised several named sections, whereas Nicolle (2015) included the same species in a single section, section Eucalyptus, divided into several series, most of which correspond with Brooker’s groupings. The analyses presented here suggest that some of these groupings should be revised. In the case of E. deuaensis, both Brooker and Nicolle placed this taxon in a series separate from series Pachyphloiae (Appendix 5), the stringybarks, but the MP, Bayesian and PADD trees clearly placed E. deuaensis within the stringybark group as sister to E. caliginosa. The series Psathyroxylon is supported as monophyletic if the monotypic series, series Stenostomae, is included. In all analyses, the sole species in this series, E. stenostoma, is consistently strongly associated (>99% BS, PP: 1) with some species of the subseries Considenianae. The position of E. consideniana itself is problematic, with the different analyses suggesting divergent affinities for the two accessions included, possibly as a result of gene flow from other species in subgenus Eucalyptus. The scribbly gums (E. haemastoma, E. sclerophylla and E. rossii), subseries Haemastomae, are well supported as monophyletic in the Bayesian analysis (PP: 1), with E. rossii indicated as sister to the other species (E. rossii is also sister to the other scribbly gums in the MP analysis, although not in the PADD tree).
All analyses indicated that series Strictae, as recognised by both Brooker (2000) and Nicolle (2015), is not monophyletic and that the rank of the two included subseries should be revised because only subseries Irregulares sensu Brooker is monophyletic, whereas subseries Regulares is paraphyletic and not unambiguously sister to subseries Irregulares. The placement of E. cunninghamii differs between their classifications; both included it within series Strictae (Appendix 5), Nicolle included it in subseries Irregulares with E. stricta and its allies, whereas Brooker placed it in subseries Regulares with E. approximans and allied species. In the present study, all analyses (>70% BS, PP: 1) agreed with Brooker’s placement. A member of Nicolle’s subseries Regulares is E. kybeanensis, which Brooker considered to be a member of the monotypic series Contiguae. Here, also, the phylogeny supports Brooker’s position; the three accessions of E. kybeanensis form a well supported clade sister to the peppermint, E. radiata, rather than to other taxa from subseries Regulares. A fourth species, E. paliformis, is included by both Brooker and Nicolle in series Strictae subseries Regulares. In this case, the MP and Bayesian analyses suggested that this species is sister to a clade that includes not only other members of subseries Regulares, but also the peppermint, E. radiata, and the black sallies. A fifth species, E. spectatrix, was not recognised by Brooker, but it was included in series Strictae subseries Irregulares by Nicolle. In all of our analyses, E. spectatrix received strong support as a distinct species, even though most other species, with the possible exception of E. langleyi (monophyletic in the Bayesian tree, but not in the MP and PADD analyses) and E. burgessiana (monophyletic in the Bayesian and PADD trees, but not in the MP analysis), did not appear monophyletic. Eucalyptus langleyi and other taxa from the Sydney region and GBMWHA are the focus of ongoing research (S. Rutherford, P. G. Wilson, M. Rossetto and S. P. Bonser, unpubl. data).
Conclusions
Phylogenetic analysis of DArT markers recovered trees that were consistent with previous phylogenies of subgenus Eucalyptus based on sequence data, with many relationships supporting those from previous classifications. However, some relationships, particularly of taxa in the Sydney region and GBMWHA, were not consistent with previous classifications, highlighting the need for a revision of the green ashes and other taxa in subgenus Eucalyptus. As with many eucalypts, relationships in the green ashes have been defined on the basis of quantitative characters such as leaf length, leaf width, fruit size and bud size, as well as geographic location. However, the results here suggest that some morphological traits may not necessarily be reflective of evolutionary relationships within and among taxa. Defining species boundaries on the basis of geographic location is likely to be equally problematic. A detailed population genomic study focussing on taxa from the Sydney region and GBMWHA is required to better understand patterns of gene flow, species boundaries and the evolutionary history of the group.
Acknowledgements
We are very grateful to our colleagues at the Royal Botanic Garden Sydney (RBG) for assistance with the collections, including Doug Benson, Andrew Orme, Bob Coveny, Michael Elgey and Trevor Wilson; and with the DNA extractions, including Carolyn Connelly, Margaret Heslewood, Juelian Siow, Marlien van der Merwe and Hannah McPherson. We also thank Tracy Armstrong (Australian Botanic Garden, Mount Annan) and Rusty Worsman (formerly Blue Mountains Botanic Garden, Mount Tomah) who facilitated access to cultivated plants; and RBG volunteers, Aaron Smith for assistance in the field and Danca Ciric for providing a specimen of E. luehmanniana. We are grateful to Dean Nicolle for providing leaf material of E. regnans, as well as Jason Carling, Cina Vipin, Vanessa Caig and Andrzej Kilian from Diversity Arrays Technology Pty Ltd who gave advice and technical support for a modified CTAB extraction for DArTs. We also thank Dorothy Steane (University of Tasmania) who provided advice on various aspects of this research, particularly with the analysis of DArT markers. Collecting in New South Wales operated under the Royal Botanic Gardens and Domain Trust (New South Wales). Collections in Snowy River and Alpine National Park, Victoria, and in Girraween National Park, Queensland, operated under Scientific Licence numbers 10006635 and WITK12361813 respectively. This research was funded by ARC Linkage Grant LP110100721. S. Rutherford is in receipt of an Australian Postgraduate Award. We also thank three anonymous reviewers for providing comments that improved the manuscript.
References
Ackerly DD, Dudley SA, Sultan SE, Schmitt J, Coleman JS, Randall Linder C, Sandquist DR, Geber MA, Evans AS, Dawson TE, Lechowicz MJ (2000) The evolution of plant ecophysiological traits: recent advances and future directions. Bioscience 50, 979–995.| The evolution of plant ecophysiological traits: recent advances and future directions.Crossref | GoogleScholarGoogle Scholar |
Atlas of Living Australia (2015) Spatial portal. Available at http://spatial.ala.org.au/?q=qid%3A1423528839339&qc=data_hub_uid:dh2# [Verified 1 August 2015]
Australia’s Virtual Herbarium (2015) Occurrence records. Available at http://avh.ala.org.au/occurrences/search?q=collector_text%3ARutherford+matched_name_children%3AEucalyptus#tab_mapView [Verified 1 August 2015]
Bayly MJ, Ladiges PY (2007) Divergent paralogues of ribosomal DNA in eucalypts (Myrtaceae). Molecular Phylogenetics and Evolution 44, 346–356.
| Divergent paralogues of ribosomal DNA in eucalypts (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmtVOltbo%3D&md5=3a6923ab5deb7de6d181ddc99cae8fe3CAS | 17188000PubMed |
Bayly MJ, Udovicic F, Gibbs AK, Parra-O C, Ladiges PY (2008) Ribosomal DNA pseudogenes are widespread in the eucalypt group (Myrtaceae): implications for phylogenetic analysis. Cladistics 24, 131–146.
| Ribosomal DNA pseudogenes are widespread in the eucalypt group (Myrtaceae): implications for phylogenetic analysis.Crossref | GoogleScholarGoogle Scholar |
Bayly MJ, Rigault P, Spokevicius A, Ladiges PY, Ades PK, Anderson C, Bossinger G, Merchant A, Udovicic F, Woodrow IE, Tibbits J (2013) Chloroplast genome analysis of Australian eucalypts: Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Molecular Phylogenetics and Evolution 69, 704–716.
| Chloroplast genome analysis of Australian eucalypts: Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXht1eltLrO&md5=924978c102a3a143cada25ffde2d8867CAS | 23876290PubMed |
Benson D, McDougall L (1998) Ecology of Sydney plant species Part 6: dicotyledon family Myrtaceae. Cunninghamia 5, 808–987.
Brooker MIH (2000) A new classification of the genus Eucalyptus L’Hér. (Myrtaceae). Australian Systematic Botany 13, 79–148.
| A new classification of the genus Eucalyptus L’Hér. (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Brooker MIH, Kleinig DA (2006) ‘Field Guide to Eucalypts’, 3rd edn. (Bloomings Books: Melbourne)
Byrne M (2007) Phylogeography provides an evolutionary context for the conservation of a diverse and ancient flora. Australian Journal of Botany 55, 316–325.
| Phylogeography provides an evolutionary context for the conservation of a diverse and ancient flora.Crossref | GoogleScholarGoogle Scholar |
Chappill JA, Ladiges PY (1996) Phylogenetic analysis of Eucalyptus informal subgenus Symphyomyrtus section Maidenaria. Australian Systematic Botany 9, 71–93.
| Phylogenetic analysis of Eucalyptus informal subgenus Symphyomyrtus section Maidenaria.Crossref | GoogleScholarGoogle Scholar |
Crisp MD, Burrows GE, Cook LG, Thornhill AH, Bowman DMJS (2011) Flammable biomes dominated by eucalypts originated at the Cretaceous–Palaeogene boundary. Nature Communications 2, 193
| Flammable biomes dominated by eucalypts originated at the Cretaceous–Palaeogene boundary.Crossref | GoogleScholarGoogle Scholar | 21326225PubMed |
Desper R, Gascuel O (2002) Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. Journal of Computational Biology 9, 687–705.
| Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XpslClurs%3D&md5=b2f695e6eedd9a07553ae6ab6ce70d35CAS | 12487758PubMed |
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12, 13–15.
Eldridge K, Davidson J, Harwood C, Van Wyk G (1993) ‘Eucalypt Domestication and Breeding.’ (Oxford University Press: New York)
Farris J (1977) Phylogenetic analysis under Dollo’s law. Systematic Biology 26, 77–88.
| Phylogenetic analysis under Dollo’s law.Crossref | GoogleScholarGoogle Scholar |
Field DL, Ayre DJ, Whelan RJ, Young AG (2011a) Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregata and common E. rubida. Heredity 106, 841–853.
| Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregata and common E. rubida.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3MvkslSqtw%3D%3D&md5=aad38881acc797bfd173bfa78ce874bdCAS | 21063438PubMed |
Field DL, Ayre DJ, Whelan RJ, Young AG (2011b) The importance of pre-mating barriers and the local demographic context for contemporary mating patterns in hybrid zones of Eucalyptus aggregata and Eucalyptus rubida. Molecular Ecology 20, 2367–2379.
| The importance of pre-mating barriers and the local demographic context for contemporary mating patterns in hybrid zones of Eucalyptus aggregata and Eucalyptus rubida.Crossref | GoogleScholarGoogle Scholar | 21375638PubMed |
Francis AR, Steel M (2015) Tree-like reticulation networks: when do tree-like distances also support reticulate evolution? Mathematical Biosciences 259, 12–19.
| Tree-like reticulation networks: when do tree-like distances also support reticulate evolution?Crossref | GoogleScholarGoogle Scholar | 25447812PubMed |
Gandolfo MA, Hermsen EJ, Zamaloa MC, Nixon KC, Gonzalez CC, Wilf P, Cuneo NR, Johnson KR (2011) Oldest known Eucalyptus macrofossils are from South America. PLoS One 6, e21084
| Oldest known Eucalyptus macrofossils are from South America.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXot1yqtLg%3D&md5=2ab7d1a4c5973728b8cc6b0866482f79CAS | 21738605PubMed |
Garland T, Bennett AF, Rezende EL (2005) Phylogenetic approaches in comparative physiology. The Journal of Experimental Biology 208, 3015–3035.
| Phylogenetic approaches in comparative physiology.Crossref | GoogleScholarGoogle Scholar | 16081601PubMed |
Govindarajulu R, Parks M, Tennessen JA, Liston A, Ashman T-L (2015) Comparison of nuclear, plastid, and mitochondrial phylogenies and the origin of wild octoploid strawberry species. American Journal of Botany 102, 544–554.
| Comparison of nuclear, plastid, and mitochondrial phylogenies and the origin of wild octoploid strawberry species.Crossref | GoogleScholarGoogle Scholar | 25878088PubMed |
Grattapaglia D, Vaillancourt RE, Shepherd M, Thumma BR, Foley W, Külheim C, Potts BM, Myburg AA (2012) Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus. Tree Genetics & Genomes 8, 463–508.
| Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus.Crossref | GoogleScholarGoogle Scholar |
Griffin AR, Burgess IP, Wolf L (1988) Patterns of natural and manipulated hybridization in the genus Eucalyptus L’Herit: a review. Australian Journal of Botany 36, 41–66.
| Patterns of natural and manipulated hybridization in the genus Eucalyptus L’Herit: a review.Crossref | GoogleScholarGoogle Scholar |
Hager T, Benson D (2010) The eucalypts of the Greater Blue Mountains World Heritage Area: distribution, classification and habitats of the species of Eucalyptus, Angophora and Corymbia (family Myrtaceae) recorded in its eight conservation reserves. Cunninghamia 11, 425–444.
Hermsen EJ, Gandolfo MA, del Carmen Zamaloa M (2012) The fossil record of Eucalyptus in Patagonia. American Journal of Botany 99, 1356–1374.
| The fossil record of Eucalyptus in Patagonia.Crossref | GoogleScholarGoogle Scholar | 22859652PubMed |
Hill KD (1991) Myrtaceae: Eucalyptus. In ‘Flora of New South Wales. Vol. 2’. (Ed. GJ Harden) pp.76–142. (New South Wales University Press: Sydney)
Hill RS (1994) Chapter 16: the history of selected Australian taxa. In ‘History of the Australian Vegetation: Cretaceous to Recent’. (Ed. RS Hill) pp. 390–419. (Cambridge University Press: Cambridge, UK).
Hill KD (2002) Myrtaceae: Eucalyptus. In ‘Flora of New South Wales. Vol. 2’, revised edn. (Ed. GJ Harden) pp. 96–164. (University of New South Wales Press: Sydney)
Hill KD, Johnson LAS (1995) Systematic studies in the eucalypts. 7. A revision of the bloodwoods, genus Corymbia (Myrtaceae). Telopea 6, 185–504.
| Systematic studies in the eucalypts. 7. A revision of the bloodwoods, genus Corymbia (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Hodson MJ, White PJ, Broadley MR (2005) Phylogenetic variation in the silicon composition of plants. Annals of Botany 96, 1027–1046.
| Phylogenetic variation in the silicon composition of plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtlSjsL3F&md5=2abdaec455cc8b2e8df1716335b9bcdaCAS | 16176944PubMed |
Hovenden MJ, Vander Schoor JK (2004) Nature vs nurture in the leaf morphology of Southern beech, Nothofagus cunninghamii (Nothofagaceae). New Phytologist 161, 585–594.
| Nature vs nurture in the leaf morphology of Southern beech, Nothofagus cunninghamii (Nothofagaceae).Crossref | GoogleScholarGoogle Scholar |
Hudson CJ, Freeman JS, Kullan ARK, Petroli CD, Sansaloni CP, Kilian A, Detering F, Grattapaglia D, Potts BM, Myburg AA, Vaillancourt RE (2012) A reference linkage map for eucalypts. BMC Genomics 13, 240
| A reference linkage map for eucalypts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhslamu7zM&md5=26a6d18ac3e612608f4ac465e04243c9CAS | 22702473PubMed |
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755.
| MRBAYES: Bayesian inference of phylogeny.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3MvotV2isw%3D%3D&md5=b8644e64d679ca4a9de09f610a1cf0f1CAS | 11524383PubMed |
Huson DH (1998) SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14, 68–73.
| SplitsTree: analyzing and visualizing evolutionary data.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXisFygs70%3D&md5=5082b5b584070d21818863580a389b7aCAS | 9520503PubMed |
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23, 254–267.
| Application of phylogenetic networks in evolutionary studies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XntValsw%3D%3D&md5=e56e55f9f49efd5f4815ee43baf3672eCAS | 16221896PubMed |
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Research 29, 25e
| Diversity arrays: a solid state technology for sequence information independent genotyping.Crossref | GoogleScholarGoogle Scholar |
Kim S-T, Donoghue MJ (2008) Incongruence between cpDNA and nrITS trees indicates extensive hybridisation within Eupersicaria (Polygonaceae). American Journal of Botany 95, 1122–1135.
| Incongruence between cpDNA and nrITS trees indicates extensive hybridisation within Eupersicaria (Polygonaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFygs73F&md5=6b18740bafe5a922e2ed20061de104d5CAS | 21632431PubMed |
Klaphake V (2012) ‘Eucalypts of the Sydney Region’, 3rd edn. (Van Klaphake: Byabarra, NSW, Australia)
Kullan ARK, van Dyk MM, Jones N, Kanzler A, Bayley A, Myburg AA (2012) High-density genetic linkage maps with over 2400 sequence-anchored DArT markers for genetic dissection in an F2 pseudo-backcross of Eucalyptus grandis × E. urophylla. Tree Genetics & Genomes 8, 163–175.
| High-density genetic linkage maps with over 2400 sequence-anchored DArT markers for genetic dissection in an F2 pseudo-backcross of Eucalyptus grandis × E. urophylla.Crossref | GoogleScholarGoogle Scholar |
Ladiges PY, Humphries CJ, Brooker MIH (1987) Cladistic and biogeographic analysis of Western Australian species of Eucalyptus L’Hérit., informal subgenus Monocalyptus Pryor & Johnson. Australian Journal of Botany 35, 251–281.
| Cladistic and biogeographic analysis of Western Australian species of Eucalyptus L’Hérit., informal subgenus Monocalyptus Pryor & Johnson.Crossref | GoogleScholarGoogle Scholar |
Ladiges PY, Newnham MR, Humphries CJ (1989) Systematics and biogeography of the Australian ‘green ash’ eucalypts (Monocalyptus). Cladistics 5, 345–364.
| Systematics and biogeography of the Australian ‘green ash’ eucalypts (Monocalyptus).Crossref | GoogleScholarGoogle Scholar |
Ladiges PY, Udovicic F, Drinnan AN (1995) Eucalypt phylogeny: molecules and morphology. Australian Systematic Botany 8, 483–497.
| Eucalypt phylogeny: molecules and morphology.Crossref | GoogleScholarGoogle Scholar |
Ladiges PY, Bayly MJ, Nelson GJ (2010) East–west continental vicariance in Eucalyptus subgenus Eucalyptus. In ‘Beyond Cladistics: the Branching of a Paradigm’. (Eds DM Williams, S Knapp) pp. 267–301. (University of California Press: Los Angeles, CA)
Lassak EV, Southwell IA (1982) The stem volatile leaf oils of some species of Eucalyptus subseries Strictinae. Phytochemistry 21, 2257–2261.
| The stem volatile leaf oils of some species of Eucalyptus subseries Strictinae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3sXhsVSms7c%3D&md5=c3247aa715c836142071ada93499a94eCAS |
Le Quesne WJ (1974) The uniquely evolved character concept and its cladistic application. Systematic Biology 23, 513–517.
| The uniquely evolved character concept and its cladistic application.Crossref | GoogleScholarGoogle Scholar |
Lefort V, Desper R, Gascuel O (2015) FastME 2.0: a comprehensive, accurate and fast distance-based phylogeny inference program. Molecular Biology and Evolution
| FastME 2.0: a comprehensive, accurate and fast distance-based phylogeny inference program.Crossref | GoogleScholarGoogle Scholar | 26130081PubMed |
Lexer C, Widmer A (2008) The genic view of plant speciation: recent progress and emerging questions. Philosophical Transactions of the Royal Society of London – B. Biological Sciences 363, 3023–3036.
| The genic view of plant speciation: recent progress and emerging questions.Crossref | GoogleScholarGoogle Scholar | 18579476PubMed |
Macphail M (2007) Australian Palaeoclimates: Cretaceous to Tertiary: a review of palaeobotanical and related evidence to the year 2000. CRC LEME special volume, open file report 151. CRC LEME, Perth, WA, Australia.
Mallet J (2005) Hybridization as an invasion of the genome. Trends in Ecology & Evolution 20, 229–237.
| Hybridization as an invasion of the genome.Crossref | GoogleScholarGoogle Scholar |
McGowen MH, Wiltshire RJE, Potts BM, Vaillancourt RE (2001) The origin of Eucalyptus vernicosa, a unique shrub eucalypt. Biological Journal of the Linnean Society. Linnean Society of London 74, 397–405.
| The origin of Eucalyptus vernicosa, a unique shrub eucalypt.Crossref | GoogleScholarGoogle Scholar |
McKinnon GE, Steane DA, Potts BM, Vaillancourt RE (1999) Incongruence between chloroplast and species phylogenies in Eucalyptus subgenus Monocalyptus (Myrtaceae). American Journal of Botany 86, 1038–1046.
| Incongruence between chloroplast and species phylogenies in Eucalyptus subgenus Monocalyptus (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXltVKrtLY%3D&md5=17a47fe27c87dc4b94fbbd7427f8fc57CAS | 10406727PubMed |
McKinnon GE, Vaillancourt RE, Jackson HD, Potts BM (2001) Chloroplast sharing in the Tasmanian eucalypts. Evolution 55, 703–711.
| Chloroplast sharing in the Tasmanian eucalypts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXktleqtLs%3D&md5=df7ac9816e98b2085779432b5d838b2eCAS | 11392388PubMed |
McKinnon GE, Jordan GJ, Vaillancourt RE, Steane DA, Potts BM (2004) Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts. Philosophical Transactions of the Royal Society of London – B. Biological Sciences 359, 275–284.
| Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts.Crossref | GoogleScholarGoogle Scholar | 15101583PubMed |
McKinnon GE, Vaillancourt RE, Steane DA, Potts BM (2008) An AFLP marker approach to lower-level systematics in Eucalyptus (Myrtaceae). American Journal of Botany 95, 368–380.
| An AFLP marker approach to lower-level systematics in Eucalyptus (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXktlyjs7Y%3D&md5=4928844adc66a77c3c8e782d028959f3CAS | 21632361PubMed |
McKinnon GE, Smith JJ, Potts BM (2010) Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species. Molecular Ecology 19, 1367–1380.
| Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXlvVWhtrk%3D&md5=c808dbdb98c0c21a640cf877ed003e46CAS | 20298471PubMed |
Moran GF, Bell JC, Prober S (1990) The utility of isozymes in the systematics of some Australian tree groups. Australian Systematic Botany 3, 47–57.
| The utility of isozymes in the systematics of some Australian tree groups.Crossref | GoogleScholarGoogle Scholar |
Morrison DA (2014) Phylogenetic networks: a review of methods to display evolutionary history. Annual Research & Review in Biology 4, 1518–1543.
| Phylogenetic networks: a review of methods to display evolutionary history.Crossref | GoogleScholarGoogle Scholar |
Nicolle D (2015) Classification of the eucalypts (Angophora, Corymbia and Eucalyptus) Version 2. Available at http://www.dn.com.au/Classification-Of-The-Eucalypts.pdf [Verified 25 August 2015]
Ochieng JW, Henry RJ, Baverstock PR, Steane DA, Shepherd M (2007a) Nuclear ribosomal pseudogenes resolve a corroborated monophyly of the eucalypt genus Corymbia despite misleading hypotheses at functional ITS paralogs. Molecular Phylogenetics and Evolution 44, 752–764.
| Nuclear ribosomal pseudogenes resolve a corroborated monophyly of the eucalypt genus Corymbia despite misleading hypotheses at functional ITS paralogs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXnslGgsLw%3D&md5=bcaa218c6ce89d5ccfe6a78058f7204aCAS | 17570687PubMed |
Ochieng JW, Steane DA, Ladiges PY, Baverstock PR, Henry RJ, Shepherd M (2007b) Microsatellites retain phylogenetic signals across genera in eucalypts (Myrtaceae). Genetics and Molecular Biology 30, 1125–1134.
| Microsatellites retain phylogenetic signals across genera in eucalypts (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFKrtbg%3D&md5=0642d7c1f0b18495fb548b42633c2b10CAS |
Parra-O C, Bayly M, Udovicic F, Ladiges P (2006) ETS sequences support the monophyly of the eucalypt genus Corymbia (Myrtaceae). Taxon 55, 653–663.
| ETS sequences support the monophyly of the eucalypt genus Corymbia (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Parra-O C, Bayly MJ, Drinnan A, Udovicic F, Ladiges P (2009) Phylogeny, major clades and infrageneric classification of Corymbia (Myrtaceae), based on nuclear ribosomal DNA and morphology. Australian Systematic Botany 22, 384–399.
| Phylogeny, major clades and infrageneric classification of Corymbia (Myrtaceae), based on nuclear ribosomal DNA and morphology.Crossref | GoogleScholarGoogle Scholar |
Pole MS, Hill RS, Green N, Macphail MK (1993) The Oligocene Berwick Quarry flora: rainforest in a drying environment. Australian Systematic Botany 6, 399–427.
| The Oligocene Berwick Quarry flora: rainforest in a drying environment.Crossref | GoogleScholarGoogle Scholar |
Pollock LJ, Bayly MJ, Nevill PG, Vesk PA (2013) Chloroplast DNA diversity associated with protected slopes and valleys for hybridizing Eucalyptus species on isolated ranges in south-eastern Australia. Journal of Biogeography 40, 155–167.
| Chloroplast DNA diversity associated with protected slopes and valleys for hybridizing Eucalyptus species on isolated ranges in south-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Pollock LJ, Bayly MJ, Vesk PA (2015) The roles of ecological and evolutionary processes in plant community assembly: the environment, hybridization, and introgression influence co-occurrence of Eucalyptus. American Naturalist 185, 784–796.
| The roles of ecological and evolutionary processes in plant community assembly: the environment, hybridization, and introgression influence co-occurrence of Eucalyptus.Crossref | GoogleScholarGoogle Scholar | 25996863PubMed |
Potts BM, Pederick LA (2000) Morphology, phylogeny, origin, distribution and genetic diversity of eucalypts. In ‘Diseases and Pathogens of Eucalypts’. (Eds PJ Keane, GA Kile, FD Podger, BN Brown) pp. 11–34. (CSIRO Publishing: Melbourne)
Potts BM, Wiltshire RJE (1997) Eucalypt genetics and genecology. In ‘Eucalypt Ecology’. (Eds JE Williams, JCZ Woinarski) pp. 56–91. (Cambridge University Press: Cambridge, UK)
Prober S, Bell JC, Moran G (1990) A phylogenetic and allozyme approach to understanding rarity in three ‘green ash’ eucalypts (Myrtaceae). Plant Systematics and Evolution 172, 99–118.
| A phylogenetic and allozyme approach to understanding rarity in three ‘green ash’ eucalypts (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXhs1Sruro%3D&md5=e7e948b8c1dd89b3860d2208b54b842aCAS |
Pryor LD, Johnson LAS (1971) ‘A Classification of the Eucalypts.’ (The Australian National University: Canberra)
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574.
| MrBayes 3: Bayesian phylogenetic inference under mixed models.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXntlKms7k%3D&md5=ebb7f79ca3abc332a555ac1fa9492884CAS | 12912839PubMed |
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542.
| MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space.Crossref | GoogleScholarGoogle Scholar | 22357727PubMed |
Rossetto M, Lucarotti F, Hopper SD, Dixon KW (1997) DNA fingerprinting of Eucalyptus graniticola: a critically endangered relict species or a rare hybrid? Heredity 79, 310–318.
| DNA fingerprinting of Eucalyptus graniticola: a critically endangered relict species or a rare hybrid?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXmtl2ltbk%3D&md5=f04b1c10e570da2aa91acff473c6d695CAS |
Rossetto M, Harriss FCL, Mclauchlan A, Henry RJ, Baverstock PR, Lee LS (2000) Interspecific amplification of tea tree (Melaleuca alternifolia – Myrtaceae) microsatellite loci: potential implications for conservation studies. Australian Journal of Botany 48, 367–373.
| Interspecific amplification of tea tree (Melaleuca alternifolia – Myrtaceae) microsatellite loci: potential implications for conservation studies.Crossref | GoogleScholarGoogle Scholar |
Rozefelds AC (1996) Eucalyptus phylogeny and history: a brief summary. Tasforests 8, 15–26.
Sale MM, Potts BM, West AK, Reid JB (1993) Relationships within Eucalyptus using chloroplast DNA. Australian Systematic Botany 6, 127–138.
| Relationships within Eucalyptus using chloroplast DNA.Crossref | GoogleScholarGoogle Scholar |
Sansaloni CP, Petroli CD, Carling J, Hudson CJ, Steane DA, Myburg AA, Grattapaglia D, Vaillancourt RE, Kilian A (2010) A high-density Diversity Arrays Technology (DArT) microarray for genome-wide genotyping in Eucalyptus. Plant Methods 6, 16
| A high-density Diversity Arrays Technology (DArT) microarray for genome-wide genotyping in Eucalyptus.Crossref | GoogleScholarGoogle Scholar | 20587069PubMed |
Semagn K, Bjørnstad Å, Ndjiondjop MN (2006) An overview of molecular marker methods for plants. African Journal of Biotechnology 5, 2540–2568.
Slee AV, Brooker MIH, Duffy SM, West JG (2006) ‘EUCLID – Eucalypts of Australia’, 3rd edn. (CD-ROM) (CSIRO Publishing: Melbourne)
Smith S, Hughes J, Wardell-Johnson G (2003) High population differentiation and extensive clonality in a rare mallee eucalypt: Eucalyptus curtisii. Conservation Genetics 4, 289–300.
| High population differentiation and extensive clonality in a rare mallee eucalypt: Eucalyptus curtisii.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXktlagsLk%3D&md5=3bb237104115b28a75d638460ed0ac68CAS |
Soltis DE, Kuzoff RK (1995) Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae). Evolution 49, 727–742.
| Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae).Crossref | GoogleScholarGoogle Scholar |
Steane DA, Byrne M, Vaillancourt RE, Potts BM (1998) Chloroplast DNA polymorphism signals complex interspecific interactions in Eucalyptus (Myrtaceae). Australian Systematic Botany 11, 25–40.
| Chloroplast DNA polymorphism signals complex interspecific interactions in Eucalyptus (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Steane DA, McKinnon GE, Vaillancourt RE, Potts BM (1999) ITS sequence data resolve higher level relationships among the eucalypts. Molecular Phylogenetics and Evolution 12, 215–223.
| ITS sequence data resolve higher level relationships among the eucalypts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXjvFWrsbk%3D&md5=574381dedb74ecead617ecba3ce0a392CAS | 10381324PubMed |
Steane DA, Nicolle D, McKinnon GE, Vaillancourt RE, Potts BM (2002) Higher-level relationships among the eucalypts are resolved by ITS-sequence data. Australian Systematic Botany 15, 49–62.
| Higher-level relationships among the eucalypts are resolved by ITS-sequence data.Crossref | GoogleScholarGoogle Scholar |
Steane DA, Nicolle D, Sansaloni CP, Petroli CD, Carling J, Kilian A, Myburg AA, Grattapaglia D, Vaillancourt RE (2011) Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping. Molecular Phylogenetics and Evolution 59, 206–224.
| Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping.Crossref | GoogleScholarGoogle Scholar | 21310251PubMed |
Thornhill AH, Macphail M (2012) Fossil myrtaceous pollen as evidence for the evolutionary history of Myrtaceae: a review of fossil Myrtaceidites species. Review of Palaeobotany and Palynology 176–177, 1–23.
| Fossil myrtaceous pollen as evidence for the evolutionary history of Myrtaceae: a review of fossil Myrtaceidites species.Crossref | GoogleScholarGoogle Scholar |
Turak E, Marchant R, Barmuta LA, Davis J, Choy S, Metzeling L (2011) River conservation in a changing world: invertebrate diversity and spatial prioritisation in south-eastern coastal Australia. Marine and Freshwater Research 62, 300–311.
| River conservation in a changing world: invertebrate diversity and spatial prioritisation in south-eastern coastal Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjsVKktr4%3D&md5=9c5b0249cd84cd8487d5df1dad3e59feCAS |
Udovicic F, Ladiges PY (2000) Informativeness of nuclear and chloroplast DNA regions and the phylogeny of the eucalypts and related genera (Myrtaceae). Kew Bulletin 55, 633–645.
| Informativeness of nuclear and chloroplast DNA regions and the phylogeny of the eucalypts and related genera (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Udovicic F, McFadden GI, Ladiges PY (1995) Phylogeny of Eucalyptus and Angophora based on 5S rDNA spacer sequence data. Molecular Phylogenetics and Evolution 4, 247–256.
| Phylogeny of Eucalyptus and Angophora based on 5S rDNA spacer sequence data.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXptFShtbY%3D&md5=e53c8464273ed00bc9345ec6a4c696e0CAS | 8845962PubMed |
Wang H, Sun D, Sun G (2011) Molecular phylogeny of diploid Hordeum species and incongruence between chloroplast and nuclear datasets. Genome 54, 986–992.
| Molecular phylogeny of diploid Hordeum species and incongruence between chloroplast and nuclear datasets.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XmsFCrsQ%3D%3D&md5=3a353d496b79ad977922276ff4fd5efdCAS | 22085287PubMed |
Whittock S, Steane DA, Vaillancourt RE, Potts BM (2003) Molecular evidence shows that the tropical boxes (Eucalyptus subgenus Minutifructus) are over-ranked. Transactions of the Royal Society of South Australia 127, 27–32.
Wilson PG (2011) Myrtaceae. In ‘The Families and Genera of Vascular Plants. Vol. 10. Flowering Plants Eudicots’. (Ed. K Kubitzki) pp. 212–271. (Springer-Verlag: Berlin)
Woodhams M, Steane DA, Jones RC, Nicolle D, Moulton V, Holland BR (2013) Novel distances for Dollo data. Systematic Biology 62, 62–77.
| Novel distances for Dollo data.Crossref | GoogleScholarGoogle Scholar | 22914977PubMed |
Wu C-I (2001) The genic view of the process of speciation. Journal of Evolutionary Biology 14, 851–865.
| The genic view of the process of speciation.Crossref | GoogleScholarGoogle Scholar |
Yeoh SH, Ho SYW, Thornhill AH, Foley WJ (2013) Regional population expansion in Eucalyptus globulus. Molecular Phylogenetics and Evolution 68, 498–501.
| Regional population expansion in Eucalyptus globulus.Crossref | GoogleScholarGoogle Scholar | 23643971PubMed |
Yu W-B, Huang P-H, Li D-Z, Wang H (2013) Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae). PLoS One 8, e74828
| Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsFeiurnJ&md5=533586351efe715f993b17e332043882CAS | 24069353PubMed |
A AAuthors of plant names are given in Table 2 and authors of both species and higher taxonomic ranks are listed in Appendices 1 and 5.
Appendix 3. Phylogeny derived from 1780 Diversity Arrays Technology (DArT) markers analysed with Partitioned Additive Dollo Distance (PADD) and minimum-evolution tree estimation of the green ashes (section Eucalyptus) and other taxa in subgenus Eucalyptus
Includes the black sallies (section Longitudinales), blue ashes (including the scribbly gums, section Cineraceae), peppermints (section Aromatica) and stringybarks (section Capillulus). Eucalyptus cloeziana (subgenus Idiogenes) is the outgroup. Sample codes correspond to those in Table 2 (Column 6). Series and subseries (Brooker 2000) within the green ashes are shown: Regnantes (R), Eucalyptus (E), Strictae subseries Irregulares (SI), Strictae subseries Regulares (SR) and Contiguae (C). Node numbers represent bootstrap values greater than 50%
Appendix 4. Network generated in SplitsTree4 (version 4.13.1) based on 2702 Diversity Arrays Technology (DArT) markers
Relationships among the green ashes (section Eucalyptus) and other taxa in subgenus Eucalyptus (sections Longitudinales, Cineraceae, Aromatica and Capillulus). Sample codes correspond to those in Table 2 (Column 6). Scale bar shows uncorrected P genetic distance equivalent to 0.01