Sensitive detection of potato viruses, PVX, PLRV and PVS, by RT-PCR in potato leaf and tuber
M. Peiman A B and C. Xie A CA Key Laboratory of Horticultural Plant Biology (Huazhong Agricultural University), Ministry of Education, National Center for Vegetable Improvement (Central China), Wuhan 430070, China.
B Takestan Islamic Azad University, Shami Shop, Talestan, Iran.
C Corresponding author. Email: xiech@mail.hzau.edu.cn
Australasian Plant Disease Notes 1(1) 41-46 https://doi.org/10.1071/DN06017
Accepted: 27 October 2006 Published: 8 November 2006
Abstract
A multiplex reverse transcription polymerase chain reaction (M-RT-PCR) was developed for the simultaneous detection of three major potato viruses, Potato virus X (PVX), Potato virus S (PVS) and Potato leaf roll virus (PLRV), in potato leaves and tubers (stored and dormant). Primers were designed from the coat protein gene of all three viruses and were able to detect virus in leaf RNA dilutions of 1 : 1024 to 1 : 4096 and from tuber RNA dilutions of 1 : 256 to 1 : 1024. The sensitivity of virus detection in leaves was higher than in tubers. All three viruses were successfully detected from tissues sampled from the tuber cortex, perimedulla and pith, with all PCR bands being strongest from cortex tissues. The reliability of the M-RT-PCR to detect PVX, PVS and PLRV in tubers was determined by testing 95 samples of field, greenhouse and in vitro potato plantlets from Wuhan, China and comparing the results with enzyme-linked immunosorbent assays (ELISA). M-RT-PCR detected all infections found by ELISA in leaf and stored tubers. In freshly harvested tubers, M-RT-PCR proved more sensitive than ELISA, as indicated by an additional 3.2% of infections detected with the M-RT-PCR method.
Additional keywords: cDNA, disease, random primer, survey.
Introduction
Potato (Solanum tuberosum) is a major food crop worldwide and is susceptible to many plant viruses including Potato virus X (Family Flexiviridae, genus Potexvirus, PVX), Potato virus S (Family Flexiviridae, genus Carlavirus, PVS) and Potato leaf roll virus (Family Luteoviridae, genus Polerovirus, PLRV). In China, the largest potato producer in the world (FAO 2006), yields are significantly reduced by 30–50% when potatoes are infected with viruses (Sun and Yang 2004). The use of certified virus-tested ‘seed’ for planting is the best way to limit the effect of these viruses on potato production (Singh 1998). The development of sensitive methods for detecting viruses is therefore of importance to support seed certification schemes (Singh and Nie 2003).
Standard methods of virus detection including mechanical inoculation to indicator hosts, electron microscopy, enzyme-linked immunosorbent assay (ELISA) and visual inspection are not adequate to reliably detect some potato viruses and might not be suitable for testing tuber samples (Mumford et al. 2000). Increased sensitivity and specificity of detection offered by reverse transcription–polymerase chain reaction (RT-PCR) is an effective method to overcome these problems (Nie and Singh 2002). PCR-based methods for PVY and PLRV detection have been shown to provide a relatively fast alternative to winter grow-out and ELISA post-harvest testing. The detection threshold of PCR-based methods was up to 128 times greater than that of an ELISA (Meunier et al. 2003).
Multiplex reverse transcription–polymerase chain reaction (M-RT-PCR) was developed for plant virus detection in 1994 (Bariana et al. 1994) with advantages of the ability to detect lower levels of virus than ELISA and the ability to discriminate two or more targets simultaneously. Recently, surveys of commercial and seed potato fields in Canada used duplex RT-PCR to detect both the common (O) and tobacco veinal necrosis strains (N/NTN) strains of PVY (Singh et al. 2003).
In this study we describe the development and optimisation of an M-RT-PCR system for the detection of PVX, PLRV and PVS from both potato leaves and tubers (freshly harvested and stored at 20–25°C for 2–4 months). The sensitivity of the M-RT-PCR was determined for different RNA concentrations and compared to ELISA, and the distribution of each virus in potato tubers was investigated.
Materials and methods
Plant material
Plants infected with PVX, PLRV and PVS were obtained from the Potato Germplasm Center of China, Potato Research Institute of Helongjiang Academy of Agricultural Sciences, China and used as positive controls. Ninety-five tuber samples randomly collected from fields and greenhouses in Wuhan, China, or were collected as microtubers sampled from in vitro plantlets (Key Laboratory of Horticultural Plant Biology, Huazhong Agricultural University). These samples were tested by M-RT-PCR and ELISA.
RNA extraction
Total RNA was extracted from potato tubers using the method of Yang et al. (2006). Tubers (300 mg) were frozen in liquid nitrogen and ground to a fine powder. To this homogenate, 1 mL of homogenisation buffer (4 M guanidine hydrochloride, 4 M urea and 1.5% thiourea) was added; the suspension was mixed and the supernatant was decanted into a tube. β-mercaptoethanol (7 µL), 1 mL ethanol (75%), 200 μL chloroform : isoamyl alcohol (24 : 1, v/v ) and 150 μL sodium acetate trihydrate (2 M, pH 4.7) were added and mixed and the tube was incubated on ice for 15 min before centrifugation for 25 min (1.01 × 104 g). The supernatant was transferred to a fresh tube, and an equal volume of pure isopropyl alcohol was added before incubation at 20°C for 1 h. Following centrifugation for 25 min (0.73 × 104g), the pellet was resuspended in 500 μL sterile DEPC-treated water and then an equal volume of phenol : chloroform : isoamyl alcohol (25 : 24 : 1, v/v/v) was added and mixed before centrifugation for 20 min (1.51 × 104g). The supernatant was transferred to a fresh microfuge tube, and an equal volume of chloroform : isoamyl alcohol (24 : 1, v/v) was added and centrifuged as described in the previous step. Sodium acetate trihydrate (100 µL) and 1 mL isopropyl alcohol were added to the supernatant in a new microfuge tube and incubated at –20°C for 15 min before centrifugation for 20 min (1.51 × 104g). The supernatant was discarded and the pellet washed with ethanol and the RNA dissolved in 25 μL of sterile DEPC-treated water (500 μL/L). Leaf RNA was isolated in the same way except that in the first step 4 M guanidine isothiocyanate, 2.5 M sodium citrate and, 0.5% sodium lauroyl sarcosine were used as the homogenisation buffer.
Reverse transcription
Complementary DNA (cDNA) was prepared by mixing 1 μL of 50 pmol random primer (6–9mer), 1 μg of total RNA, 2 μL of dNTP mixture (10 mM each), 0.5 μL of 20 U RNase inhibitor and 1 μL of reverse transcriptase (AMV) [Takara Biotechnology (Dalian) Co., Ltd] and incubated at room temperature for 10 min, and then at 42°C for 1 h, finally chilling on ice for 2 min.
Primer design and PCR
In order to design the virus-specific primers, coat protein sequences were obtained from the GenBank database for each of the viruses and were compared with CLUSTAL ‘W’ multiplex sequence alignment software (Higgins et al. 1994). For each virus one set of primers was designed (Table 1), and the properties of the primers were analysed with Primer Premier software (version 5.00, Premier Biosoft International). Biochemical optimisation of the amplification conditions for each set of primers was performed with 1 μL cDNA in a final volume of 20 μL containing 2 μL of 10× PCR buffer, 1.5 μL of 25 mM MgCl2, 2 μL of 2 mM dNTP, 0.2 μL of 5 U Taq DNA polymerase (Shanghai Promega), and 1 μL of each primer (10 μM) (forward and reverse) for RT-PCR or 0.3 μL of each primer (PLRV, PVS and PVX) for M-RT-PCR. PCR conditions were 94°C for 1.5 min (initial denaturation), 38 cycles of 94°C for 45 s (denaturation), 53°C for 45 s (primer annealing) and 72°C for 1 min (extension), followed by final extension for 5 min at 72°C. Finally, 6 μL of the products were analysed by electrophoresis on a 1.2% agarose gel.
|
ELISA
ELISA tests for PVX, PVS and PLRV were carried out following the method described by Clark and Adams (1977) using antiserum provided by Dr L. Salazar, (International Potato Center, Peru). Three replications of stolon tissue samples (1 g) from tubers both freshly harvested and stored for 2–4 months at 20–25°C after harvesting were used in ELISA tests.
Distribution of viruses in the tuber and the reliability of RT-PCR
In order to determine the distribution of virus in the tuber and the reliability of RT-PCR, RNA was extracted from three different parts of the harvested tubers: the cortex, perimedulla and pith tissue (Liu and Xie 2001). A sample of each tissue type (500 mg) was taken from each tuber using a cutter disinfected with alcohol (75% v/v) between each tuber sample. To determine the reliability of detecting virus by RT-PCR in tubers and leaves, three undiluted RNA samples for each virus were diluted 4-fold with water up to 1 : 4096 (leaves) and up to 1 : 1024 (freshly harvested tubers).
Results
Amplification of the PVX, PLRV and PVS-specific products
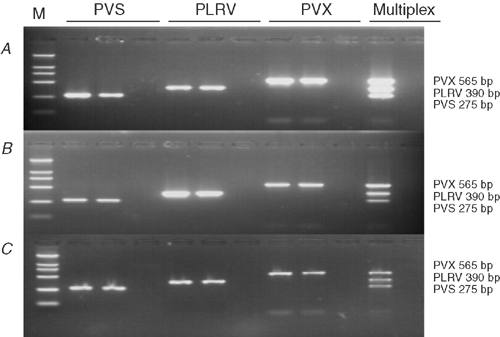
Primers designed from the coat protein gene successfully detected PVX, PLRV and PVS from both potato leaves and tubers (stored and freshly harvested; Fig. 1). No amplified product was observed with plant tissues not infected with these viruses. PVX, PLRV and PVS primer pairs, when used together in the multiplex reaction, amplified only specific products of the expected sizes, 565, 391 and 275 bp, respectively, and each amplicon was easily distinguished from the other amplicons when viewed on an agarose gel (Fig. 1).
|
M-RT-PCR sensitivity
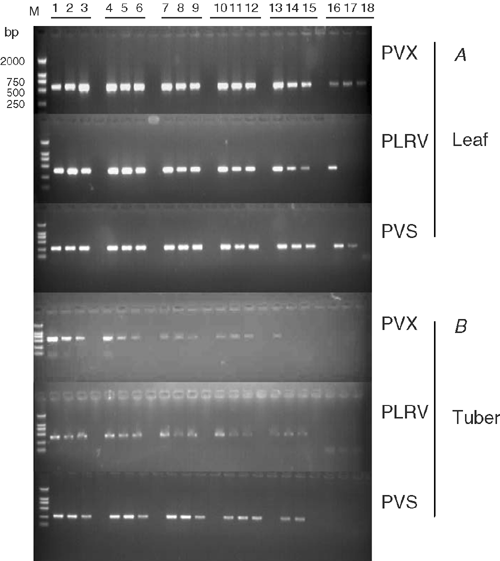
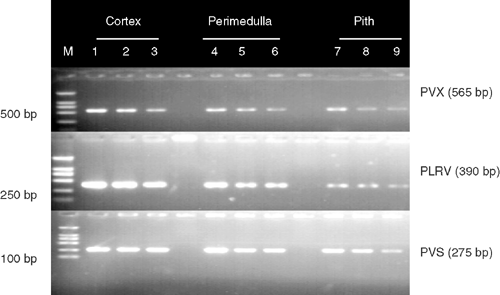
Sensitivity of M-RT-PCR was estimated by testing total RNA extracted from virus-infected potato plants diluted in 4-fold increments with water up to 1 : 4096 in leaves and up to 1 : 1024 for freshly harvested tubers. All viruses were detected in all samples of leaf dilutions up to 1 : 1024 and in 67% of leaf samples diluted up to 1 : 4096 (Fig. 2A). All three viruses were detected in all tuber samples at dilutions up to 1 : 256, and in 67% of tuber samples diluted up to 1 : 1024 (Fig. 2B). All three viruses were successfully detected from all three tuber tissue types in all replicates. Amplified bands for all three viruses were stronger from samples from the cortex tissues than from the perimedulla and pith tissues (Fig. 3, lanes 7–9).
|
|
Comparison of M-RT-PCR and ELISA
In 3.2% of the 95 tuber samples tested for PLRV, PVX and PVS by ELISA and M-RT-PCR, the PCR results differed with the visual and ELISA results, in all cases detecting additional infections (Fig. 4). This was particularly evident in samples from freshly field harvested tubers. For example, RT-PCR showed that freshly harvested tubers of potato cultivar E62 were infected with PVS and PVX but ELISA indicated these tubers were infected only with PVS in all of the three replicates. RT-PCR showed that cultivar Juan Xin Wu was infected with PVS but this infection was not evident in the ELISA results. Storage of the tissues at 25°C for 2–4 months did not affect the reliability of detection by RT-PCR.
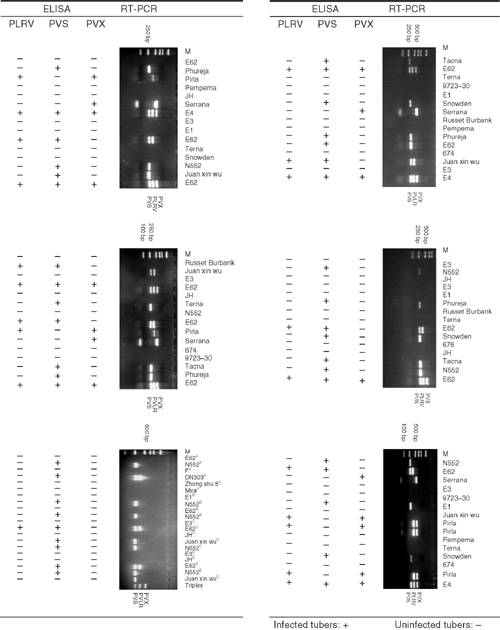
|
Discussion
PVX, PVS and PLRV are common virus diseases of potato worldwide. Infections with these pathogens may result in the loss of certification for seed crops and affect quality and yield in commercial production. Significant time and resources are spent to produce seed potato crops that contain low or undetectable levels of the virus. Potato growers in China have had increasing problems ensuring the continuity of supply of low virus seed lots.
The most common method for the detection of plant viruses is ELISA because it is rapid, inexpensive, specific and robust. However, the serological methods have the drawbacks of limited availability of antisera and questions regarding the specificity of antisera produced from preparations containing virus mixtures. Raising antisera is also time-consuming. RT-PCR has become an important method for potato pathogen detection (Petrunak et al. 1991). By contrast, a diagnostic technique using RT-PCR can be rapidly implemented in independent laboratories after the basic protocol and primer sequences are made available. However, monospecific RT-PCR assays requiring separate amplification of each virus of interest are potentially expensive and resource intensive. To overcome these limitations, there has been a keen interest in the development of new nucleic acid-based assays. M-RT-PCR has a significant advantage in that it permits simultaneous amplification of several viruses in a single reaction (Park et al. 2005; James et al. 2006). In a previous report, an RT-PCR assay, based on amplification of cDNA synthesised from genomic viral RNA, was evaluated to detect potato virus S (Heldak 2001). In this study we report the detection of PVS, PVX and PLRV in a single reaction, with the primers designed to suit an M-RT-PCR protocol.
Our research demonstrates that the coat protein genes of PVS, PVX and PLRV are reliable regions of the virus genome for the detection of these viruses using M-RT-PCR. In designing the virus-specific primers for multiplex PCR, annealing and melting temperature compatibility and the relative size of the resulting amplicon are very important (Singh and Nie 2003). Previous research has shown that shorter fragments amplify more efficiently than longer lengths (Singh and Singh 1998). In our M-RT-PCR results, cDNA from each virus was amplified to intense bands of sufficiently different sizes (Fig. 1) to allow effective separation on agarose gels. Another important factor is the quality of cDNA and we have clearly demonstrated that random primers used for making cDNA were successful for detecting these three viruses from potato tuber. Experience has shown that for expanding M-RT-PCR some reagent concentrations must be changed. In M-RT-PCR, a degenerated primer can cause significant problems in developing this method (Sharman et al. 2000). In this study, the amount of primers was reduced from 1 μL (for detecting one virus) to 0.3 μL (10 μM).
Variation in band intensity detected in different tuber tissues suggested that the cortex is the most suitable part of the tuber for detecting PVS, PVX and PLRV by M-RT-PCR, because, in the present research, it showed higher band intensity than other tissues (Fig. 3).
In conclusion, the production of virus-free seed potatoes is one of the most important steps in potato production and diagnostic assays to detect plant viruses are needed to assist the generation of high health planting material. In this study we have shown the increased sensitivity of RT-PCR over ELISA, for the detection of PLRV, PVS and PVX in potato leaves and tubers. We have also demonstrated the increased efficiency of M-RT-PCR, which can detect all three viruses individually in the one reaction.
Acknowledgements
We are very grateful to Dr Luis Salazar, International Potato Center (CIP) for his gift of antisera. Also, special thanks to him and Prof. Gad Leobenstein, Agricultural Research Organization (ARO), The Volcani Center for correcting the English and his critical reading of our manuscript. We also thank Prof. Liu Jun for her advice on technical matters. Two anonymous reviewers also made valuable comments. This work is supported by a post-doctoral fellowship from the Chinese government.
Bariana HS,
Shannon AL,
Chu PWG, Waterhouse PM
(1994) Detection of five seedborne legume viruses in one sensitive multiplex polymerase chain reaction test. Phytopathology 84, 1201–1205.
(verified 25 October 2006)
Heldak J
(2001) detection of potato viruses by RT-PCR in potato regenerants derived from in vitro heat-treated shoot tips. Acta Fytotechnica et Zootechnica 4, 275–277.

Higgins D,
Thompson J,
Gibson T,
Thompson JD,
Higgins DG, Gibson TJ
(1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680.
| PubMed |

James D,
Varga A,
Pallas V, Candresse T
(2006) Strategies for simultaneous detection of multiple plant viruses. Canadian Journal of Plant Pathology 28, 16–29.

Liu J, Xie C
(2001) Correlation of cell division and cell expansion to potato microtuber growth in vitro. Plant Cell, Tissue and Organ Culture 67, 159–164.
| Crossref | GoogleScholarGoogle Scholar |

Meunier A,
Schmit JF,
Stas A,
Kutluk N, Bragard C
(2003) Multiplex reverse transcription-PCR for simultaneous detection of beet necrotic yellow vein virus, beet soilborne virus, and beet virus Q and their vector Polymyxa betae KESKIN on sugar beet. Applied and Environmental Microbiology 69, 2356–2360.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Mumford RA,
Walsh K,
Barker L, Boonham N
(2000) Detection of potato mop top virus and tabacco rattle virus using a multiplex-real-time fluorescent reverse-transcription polymerase chain reaction assay. Phytopathology 90, 448–453.

Nie X, Singh RP
(2002) A new approach for the simultaneous differentiation of biological and geographical strains of potato virus Y by uniplex and multiplex RT-PCR. Journal of Virological Methods 104, 41–54.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Park KS,
Bae YJ,
Jung EJ, Kang SJ
(2005) RT-PCR-based detection of six garlic viruses and their phylogenetic relationships. Journal of Microbiology and Biotechnology 15, 1110–1114.

Petrunak DM,
Gildow FE, Christ BJ
(1991) Incidence and distribution of six viruses infecting potatoes in Pennsylvania. Plant Disease 75, 644.

Sharman M,
Thomas JE, Dietzgen RG
(2000) Development of a multiplex immunocapture PCR with colourimetric detection for viruses of banana. Journal of Virological Methods 89, 75–88.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Singh RP
(1998) Reverse-transcription polymerase chain reaction for the detection of viruses from plants and aphids. Journal of Virological Methods 74, 125–138.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Singh RP, Nie X
(2003) Multiplex virus and viroid detection and strain separation via multiplex reverse transcription polymerase chain reaction. Canadian Journal of Plant Pathology 25, 127–134.

Singh RP, Singh M
(1998) Specific detection of potato virus A in dormant tubers by reverse-transcription polymerase chain reaction. Plant Disease 82, 230–234.

Singh RP,
McLaren DL,
Nie X, Singh M
(2003) Possible escape of a recombinant isolate of Potato virus Y by serological indexing and methods of its detection. Plant Disease 87, 679–685.

Yang JW,
Song BT,
Li YJ, Liu J
(2006) A simple and efficient method for RNA extraction from potato tuber. Journal of Agricultural Biotechnology 14, 297–298.



