First report of a subgroup IA Cucumber mosaic virus isolate from gladiolus in India
V. K. Dubey A ,A Molecular Virology Laboratory, National Botanical Research Institute, Lucknow 226 001, India.
B M.J.P. Rohilkhand University, Bareilly, India.
C Corresponding author. Email: amin_nbri@yahoo.com.
Australasian Plant Disease Notes 3(1) 35-37 https://doi.org/10.1071/DN08014
Submitted: 17 September 2007 Accepted: 23 March 2008 Published: 4 April 2008
Abstract
This is the first report of a subgroup IA Cucumber mosaic virus isolate occurring in India. The study was based on reverse transcription-polymerase chain reaction and analysis of coat protein gene sequences.
Gladiolus is an important component of world floriculture industry and ranks among the top six flowers of export value (Anon. 1997). In recent years, a decline in gladiolus production has been observed despite of its high demand. The major factor contributing to the decline is a wide array of diseases of biological origin. Viral diseases are important because they not only cause direct damage to the host but also predispose the plants to secondary invaders (Beute 1970). Gladiolus cultivars are susceptible to many viruses, Cucumber mosaic virus (CMV) being the most prevalent (Bridgmon and Walker 1952). CMV causes severe mosaic symptoms on leaves (Fig. 1), colour break symptoms in flowers and reduced plant vigour. Vegetative propagation of gladioli results in perpetuation of viruses from one generation to another leading to a decrease in the quality and plant yield.

|
CMV is a member of the family Bromoviridae and is one of the most important wide spread viruses in the world infecting the largest number of plant species. (~1000 species). The CMV isolate occurring on gladiolus (Gladiolus psittacinus var. Hookeri cv. Red) exhibits mosaic leaves, stunted plants and colour breaking symptoms in flowers. The genome of CMV consists of three positive sense, single-stranded, RNAs (RNA 1, RNA 2 and RNA 3) and a sub genomic RNA (RNA 4) encoded by RNA 3 (Palukaitis et al. 1992) that is involved in encapsidation (Suzuki et al. 1991). Several CMV isolates reported from all over the world, have been placed into two sub groups I and II, on the basis of serology (Wahyuni et al. 1992), nucleic acid hybridisation (Owens and Palukaitis 1988) and gene sequences (Owens et al. 1990). CMV subgroup I has been recently divided into IA and IB on the basis of gene sequences available for CMV strains and phylogenetic analysis (Palukaitis and Zaitlin 1997; Roossinck et al. 1999; Roossinck 2002). Further, Asian strains of CMV have been placed into subgroup IB (Palukaitis and Zaitlin 1997). This study has been carried out to characterise CMV isolate occurring on gladiolus (CMV-Glad) using coat protein (CP) gene analysis.
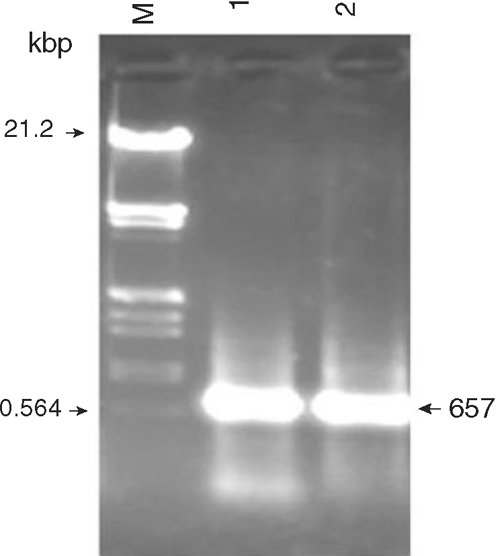
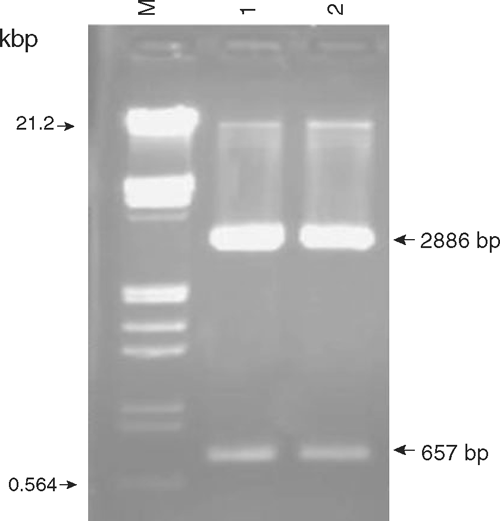
Total RNA from infected leaves of gladiolus was isolated and reverse transcription-polymerase chain reaction (RT-PCR) was performed employing CMV-CP gene specific primers (forward primer: CPF-5′-ATGGACAAATCTGAATCAAC-3′ and reverse primer: CPR-5′-CTAAACTGGGAGCACCC-3′) that amplified 657 bp product (Fig. 2). To check the identity and authenticity of PCR amplified products, Southern hybridisation was performed using α-32P labelled DNA probe prepared from the cloned CMV CP gene of a strain isolated from Catharanthus roseus (V. K. Dubey, Aminuddin and S. K. Singh, unpubl. data). Positive signals of hybridisation of the CMV probe with the amplified products confirmed the identity of the PCR amplicon as a genomic fragment derived from CMV-CP gene. The PCR product was eluted and cloned in pTZ57R/T vector (Fermentas Life Sciences). The positive transformants were screened on ampicillin, IPTG and X-gal plates and further confirmed by restriction analysis (Fig. 3).
|
|
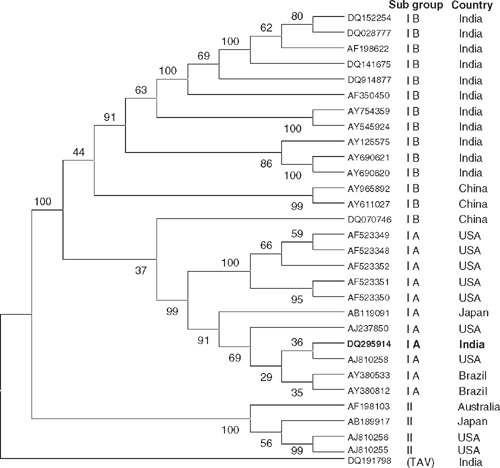
The clone was sequenced and sequencing data were submitted to GenBank (Accession number DQ295914). The BLAST (Basic Local Alignment Search Tool) search analysis of sequence data revealed highest homology with a CP gene of CMV isolate reported from USA (99% nucleotide identity). Further phylogenetic analysis of the gladiolus CMV-CP and worldwide CMV CP gene sequences was performed using multiple sequence alignment through the CLUSTAL W program. The phylogenetic tree constructed with the Mega 3.1 program (Fig. 4) shows that the CMV-Gladiolus isolate (DQ295914) belongs to subgroup IA rather than subgroup IB in contrast to other Indian isolates. This phylogeny suggests that the CMV-Gladiolus isolate did not originate from India. One interesting feature observed from the phylogenetic tree is the position of the China isolate DQ070746 that is reported to belong to subgroup IB (Dehui et al. 2006). This strain forms an intermediate position between subgroup IA and IB and may possibly represent a recombinant strain.
|
This report describes the first identification of a CMV isolate from India which shows high resemblance with subgroup IA and the USA isolate. One possible explanation is that this isolate might have introduced from the USA into India.
Acknowledgements
The authors express their gratitude to the Director, NBRI, Lucknow for providing facilities and the Council of Scientific and Industrial Research, New Delhi for financial assistance to V. K. Dubey.
Anon.
(1997) Gene bank for 150 Gladiolus varieties developed. Agriculture News 3(May, June), 139.

Beute MK
(1970) Effect of certain virus infection on susceptibility to certain fungus disease and yield of Gladiolus. Phytopathology 60, 65–70.

Bridgmon GH, Walker JC
(1952) Gladiolus as a virus reservoir. Phytopathology 42, 65–70.

Dehui X,
Liqiong L,
Jianhui W,
Weilin X,
Benchun X, Honghui L
(2006) Variation analysis of two cucumber mosaic viruses and their associated satellite RNAs from sugar beet in China. Virus Genes 33, 293–298.
| PubMed |

Owens J, Palukaitis P
(1988) Characterization of cucumber mosaic virus. I. Molecular heterogeneity mapping of RNA 3 in eight CMV strains. Virology 69, 496–502.

Owens J,
Shintaku M,
Aeschleman P,
Tahar SF, Palukaitis P
(1990) Nucleotide sequence and evolutionary relationship of Cucumber mosaic virus (CMV) strain CMV RNA 3. Journal of General Virology 71, 2243–2249.
| PubMed |

Palukaitis P, Zaitlin M
(1997) Replicase mediated resistance to plant virus diseases. Advances in Virus Research 48, 349–377.
| PubMed |

Palukaitis P,
Roossinck M,
Dietzgen RG, Francki RIB
(1992) Cucumber mosaic virus. Advances in Virus Research 41, 349–377.
| PubMed |

Roossinck MJ
(2002) Evolutionary history of cucumber mosaic virus deduced by phylogenetic analyses. Journal of Virology 76, 3382–3387.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Roossinck MJ,
Zhang L, Hellward K
(1999) Rearrangements in 5′ ontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. Journal of Virology 73, 6752–6758.
| PubMed |

Suzuki M,
Kuwata S,
Kataoka J,
Masuta C,
Nitta N, Takanami Y
(1991) Functional analysis of deletion mutants of Cucumber mosaic virus RNA 3 using an in vitro transcription system. Virology 183, 106–113.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wahyuni WS,
Dietzen RG,
Handa K, Francki RIB
(1992) Serological and biological variation between and within subgroup I and II strain of Cucumber mosaic virus. Plant Pathology 41, 282–297.
| Crossref | GoogleScholarGoogle Scholar |



