Characterisation of Apple chlorotic leaf spot virus infecting almonds in India
T. Rana A , V. Chandel A , V. Hallan A and A. A. Zaidi A BA Plant Virology Laboratory, Institute of Himalayan Bioresource Technology, Palampur 176 061, Himachal Pradesh, India.
B Corresponding author. Email: zaidi_aijaz@yahoo.com
Australasian Plant Disease Notes 3(1) 65-67 https://doi.org/10.1071/DN08026
Submitted: 5 August 2007 Accepted: 23 April 2008 Published: 5 May 2008
Abstract
Restricted field surveys were conducted to determine the presence and incidence of virus infections in almonds. Representative asymptomatic samples from six locations were tested by enzyme linked immunosorbent assay and reverse transcription and polymerase chain reaction for known stone fruit viruses. Only Apple chlorotic leaf spot virus (ACLSV) infection was detected in one of the six almond samples from the state of Himachal Pradesh. Comparison of sequenced amplicons with two available sequences of ACLSV-almond isolates revealed 92% and 94% amino acid identity. However, the Indian ACLSV-almond isolate shared maximum (98%) homology at the amino acid level to ACLSV-peach (AM498050) and wild apricot (AM498048) isolates from India. The C-terminus is highly conserved within the Indian ACLSV isolates with the exception of a few residues which in the case of the Indian almond isolates are more typical of peach and wild apricot isolate sequences.
Additional Key words: ACLSV, coat protein, RT-PCR.
Almond (Prunus amygdalus) is known as the king of nuts and is highly nutritious. It is closely related to peach and other stone fruits and is grown widely in temperate areas of the world for its edible seeds. Disease-causing agents such as bacteria, fungus and viruses result in great losses to the production of almonds. Some of the serious diseases causing heavy losses are known to be caused by viral and viroid infections (Nemeth 1986). Viruses like Apple mosaic virus (ApMV), Prune dwarf virus (PDV), Prunus necrotic ring spot virus (PNRSV) (Digiaro et al. 1992) and Plum pox virus (PPV) (Pribek et al. 2001) have been reported on almond. In one study, Apple chlorotic leaf spot virus (ACLSV) was found to be the least frequent virus infecting almonds (Barba et al. 1985).
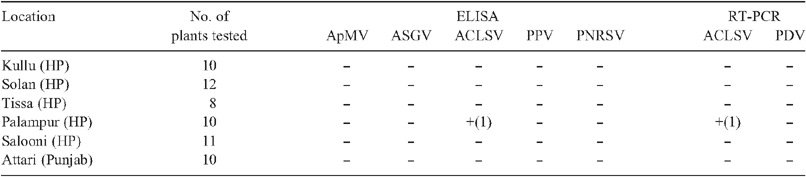
In Himachal Pradesh (HP), India, almonds are grown in mixed plantations with pome and other stone fruits and, therefore, may be exposed to the viruses that more commonly infect these plants. An initial survey was undertaken in the states of HP and Punjab to identify the major viruses infecting almonds. Almond leaf samples from Kullu, Solan, Palampur, Salooni and Tissa areas within HP and Attari within Punjab were collected. There were no visible symptoms observed and plants looked healthy.
Preliminary detection for viral infection was done by enzyme linked immunosorbent assay (ELISA) using commercially available reagents for ApMV, ACLSV, PPV (Agdia Inc., USA), PNRSV and Apple stem grooving virus (Loewe, Germany) as per the manufacturers’ instructions. ELISA readings were mostly negative except for one almond sample from HP which gave approximately twice the ELISA reading when compared with the negative control for ACLSV (Table 1). No amplification from any sample was obtained for PDV using a virus-specific primer pair (Mekuria et al. 2003) in the polymerase chain reaction (PCR).
|
Host range studies onto Chenopodium amaranticolor, C. quinoa, Phaseolus vulgaris, Nicotiana glutinosa, Cucumis sativus, N. benthamiana, and N. tabacum were conducted by mechanical inoculation using carborundum and phosphate buffer (pH 7) with 2% polyvinyl pyrrolidone (PVP) as an additive. Three replicates were conducted in which five plants of each species were inoculated for each experiment. Symptoms were observed over 12 days. C. amaranticolor and C. quinoa plants, which had been inoculated with the positive almond sample, showed small chlorotic spots in inoculated leaves and the presence of ACLSV in these plants was reconfirmed by ELISA. A single lesion from C. amaranticolor was inoculated onto the host C. quinoa, which gave chlorotic and necrotic spots on inoculated leaves, followed by chlorotic spots in upper leaves.
To further elucidate the identity of the infecting virus, total RNA was extracted using RNeasy Plant mini kit (QIAGEN, Germany) from almond leaf tissue that had tested positive in the ELISA. The primer pair (forward primer 5′-GAT CAG AAG RMG RAG GAT-3′ and reverse primer 5′-GTA GTA AAA TAT TTA AAA G-3′, accession numbers AM490253 and AM490254 respectively) was designed to amplify the complete coat protein (CP) gene and untranslated (UTR) region from the 3′ end of the ACLSV genome. Complementary DNA was synthesised by reverse transcription from 7 µL (2–4 μg), total RNA in a 25 µL reaction mixture using 200 units of Moloney Murine Leukemia Virus reverse transcriptase (M-MuLV RT) enzyme to extend from the reverse primer. PCR amplification was performed using a GeneAmp PCR 9700 machine (Applied Biosystems, USA) in a 50 µL reaction mixture with 1.5 units of Taq DNA polymerase (Invitrogen, USA) and 7 µL of the first strand cDNA reaction. The thermal cycling scheme was 2 min at 94°C, 30 cycles of 40 s at 94°C, 1 min at 49°C, 2 min at 72°C for 2 min, followed by a final incubation of 10 min at 72°C (Sambrook et al. 1989). The 800 bp amplicon was eluted using an Au-Prep gel extraction kit (Life Technologies, USA) and cloned into a pGEM-T easy vector (Promega USA). Three positive clones were sequenced in an automated sequencer (ABI Prism 310) using the Sanger dideoxy chain termination method. Three clones of the CP amplicon were sequenced twice in both directions using universal T7 and SP6 primers. The clones were identical and included 582 bp of the CP gene and 196 nt of 3′ UTR of ACLSV (GenBank accession AM498046). The sequence was also compared with partial CP sequences of ACLSV-almond isolates available from public databases (Fig. 1).
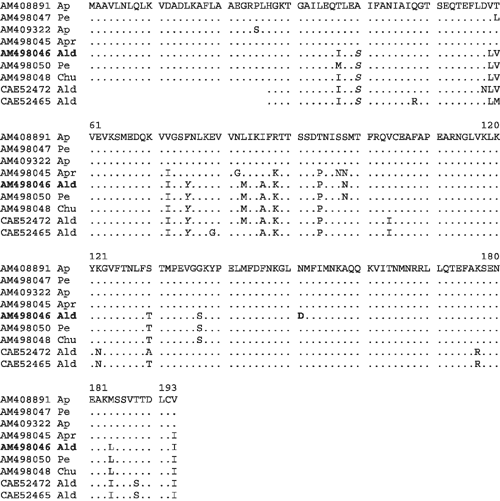
|
Al-Rwahnih et al. (2004) observed that most ACLSV-CP isolates vary only slightly in the N-terminal portion and that the C-terminal is maximally conserved. The ACLSV-CP sequence in the present study was therefore aligned with 64 other ACLSV-CP sequences (data not shown). Throughout the CP, the Indian almond isolate showed amino acid differences from all 64 ACLSV-CP sequences. The ACLSV-almond sequence from HP was most similar (98%) at the amino acid level to ACLSV-peach (AM498050) and wild apricot (AM498048) isolates from India. The two ACLSV-apple isolates (AM408891 and AM409322) and another peach isolate from HP (AM498047) had 91% sequence homology with the Indian ACLSV-almond at the amino acid level, while cultivated apricot showed 93% identity. The two Italian ACLSV-almond isolates (GenBank accessions CAE52465 and CAE52472) shared 92% and 94% identity with the Indian almond ACLSV isolate. The almond ACLSV-CP sequence was unique as it had an aspartic acid residue at position 151 instead of an asparagine residue, which was conserved in all other sequences (Fig. 1). Other important differences in the Indian almond-ACLSV isolate are serine and tyrosine at positions 40 and 75 whereas most of the other 64 isolates have alanine and phenylalanine, respectively. However, peach, wild apricot and the Italian isolates also have serine and tyrosine at positions 40 and 75. This change of alanine to serine at the 40th position and phenylalanine to tyrosine at the 75th position has been reported to result in an extreme reduction in the accumulation of viral genomic RNA, double-stranded RNAs and viral proteins (movement protein and CP) in infiltrated tissues, suggesting that alanine at position 40 and phenylalanine at position 75 are important for effective replication in host-plant cells (Yaegashi et al. 2007). This could also be responsible for reduced incidence of ACLSV in almonds in this study and reason for ACLSV being least frequent virus infecting almond (Barba et al. 1985).
This is the first molecular proof of ACLSV infection on almond in India. Wild peach (Prunus persica) and wild apricot (Prunus armeniaca), commonly called ‘Chuli’ in HP, are the two most commonly used rootstocks for almond in HP and ACLSV infection has previously been detected at the molecular level in apricot (Rana et al. 2007) and peach (AM498047 and AM498050) in HP orchards. However, the rootstock variety which might have been used for the ACLSV infected almond could not be ascertained. As ACLSV has no known vector and is transmitted through grafting, mechanical inoculation or unclean horticultural practices, the possible cause for this single infection could be graft transmission or pruning with contaminated equipment. The frequency of conserved residues shared among the Indian peach, apricot and almond isolates could be a result of grafting or mixed cropping of stone fruits.
Acknowledgements
The authors express gratitude to the Director, Institute of Himalayan Bioresource Technology, Palampur (HP), India for encouragement and providing the necessary facilities and to the Department of Science and Technology (Grant no. SR/SO/PS-71/05), Govt. of India.
Al-Rwahnih M,
Turturo C,
Minafra A,
Saldarelli P,
Myrta A,
Pallas V, Savino V
(2004) Molecular variability of Apple chlorotic leaf spot virus in different hosts and geographic regions. Journal of Plant Pathology 86, 117–122.

Barba M,
de Sanctis F, Cupidi A
(1985) Distribution of almond viruses in Central Italy. Phytopathologia Mediterranea 24, 267–269.

Digiaro M,
Savino V,
di Terlizzi B, Martelli GP
(1992) The relationship of ilarviruses to almond mosaic. Advances in Horticultural Science 6, 161–166.

Mekuria G,
Ramesha SA,
Alberts E,
Bertozzi T,
Wirthensohn M,
Collins G, Sedgley M
(2003) Comparison of ELISA and RT-PCR for the detection of Prunus necrotic ring spot virus and prune dwarf virus in almond (Prunus dulcis). Journal of Virological Methods 114, 65–69.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pribek D,
Palkovics L, Gaborjanyi R
(2001) Molecular characterization of Plum pox virus almond isolate. Acta Horticulturae 550, 91–95.

Rana T,
Chandel V,
Hallan V, Zaidi AA
(2007) Molecular evidence for Apple Chlorotic Leaf Spot Virus in wild and cultivated apricot in Himachal Pradesh, India. Journal of Plant Pathology 89, S72.

Yaegashi H,
Isogai M,
Tajima H,
Sano T, Yoshikawa N
(2007) Combinations of two amino acids (Ala40 and Phe75 or Ser40 and Tyr75) in the coat protein of apple chlorotic leaf spot virus are crucial for infectivity. Journal of General Virology 88, 2611–2618.
| Crossref | GoogleScholarGoogle Scholar | PubMed |



