Natural occurrence of Cucumber mosaic virus associated with mottling and mosaic disease on Salvia splendens, a new record from India
S. Ali A , A. A. Khan A B , B. Afreen A , R. Sharma A , T. Jahan A and Q. A. Naqvi AA Plant Virology Laboratory, Aligarh Muslim University, Aligarh-202 002, India.
B Corresponding author. Email: akil_nbri@yahoo.com
Australasian Plant Disease Notes 3(1) 118-120 https://doi.org/10.1071/DN08047
Submitted: 7 April 2008 Accepted: 4 August 2008 Published: 21 August 2008
Abstract
Natural occurrence of Cucumber mosaic virus (CMV) subgroup IB associated with mottling and mosaic symptoms on Salvia splendens was identified on the basis of serological and molecular tests for the first time in India.
Salvia (Salvia splendens Sellow ex Schult., family Lamiaceae) is an important annual ornamental plant grown worldwide. During November 2007 to March 2008, severe mottling and mosaic was observed on the leaves of S. splendens plants growing in the garden of Aligarh Muslim University, Aligarh, India, with a high disease incidence (~48.7%). Symptoms on the infected S. splendens suggest a virus as the causal pathogen.
For mechanical transmission of the virus, newly emerged leaf tissue of naturally infected S. splendens was macerated in 0.1 M phosphate buffer, pH 7.0 (1 : 1, w : v) and inoculated on several test species following the protocol of Noordam (1973). Necrotic lesions were obtained on Chenopodium amaranticolor and C. album 7–10 days post-inoculations (dpi), while systemic mosaic was observed on Nicotiana benthamiana, N. tabacum cv. White Burley and Cucumis sativus 15 dpi. To identify the virus at the serological level, direct antigen coated-enzyme linked immunosorbent assay (DAC-ELISA; Verma et al. 2005) was performed with crude sap obtained from infected leaf tissue, Cucumber mosaic virus (CMV) specific polyclonal antiserum (PVAS 242a, ATCC, USA) and alkaline phosphatase-linked secondary antibodies (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH or DSMZ, Germany). The mean absorbance at 405 nm for negative and positive controls were 0.061 ± 0.008 and 0.349 ± 0.003, respectively, while infected samples were more than four-times the value of negative controls with values that ranged between 0.289 ± 0.005 and 0.325 ± 0.003. Biological and serological identification of the virus suggests the association of CMV with the disease.
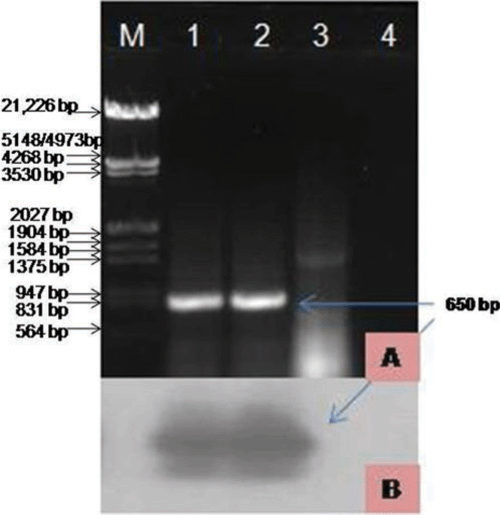
To characterise the virus at the molecular level, total RNA was isolated using RNAqueous Kit (Ambion, USA) from naturally infected and apparently healthy S. splendens leaf tissue from plants in the same location. Reverse transcription-polymerase chain reaction (RT–PCR) was performed using total RNA as templates and CMV coat protein (CP) gene specific reverse and forward primers (Srivastava et al. 2004). First strand cDNA synthesis was performed using total RNA, 50 pM reverse primers and 200 U Moloney Murine Leukemia Virus reverse transcriptase enzyme in a 20 µL reaction mixture, incubated at 42°C for 90 min. PCR was carried out in a 50 µL reaction using 5 µL cDNA as a template, 3U Taq DNA polymerase, 25 pM forward and reverse primers each, 10 mM dNTPs of each in a GeneAmp PCR 9700 system thermal cycler (Applied Biosystem, USA). The PCR conditions were as follows: initial denaturation for 3 min at 94°C followed by 30 cycles of 94°C for 15 s, 54°C for 30 s and 72°C for 30 s with a final extension at 72°C for five minutes. RT–PCR resulted in the amplification of ~650 bp expected size band from infected S. splendens, but not from asymptomatic, when assessed by gel electrophoresis (Fig. 1A). To further confirm the identity of the PCR amplicons, southern hybridisation was performed using α32P labelled DNA probe prepared from a CMV CP clone (accession EU140547) (Fig. 1B). The strong positive signals were obtained from infected samples, however, no such hybridisation signals were found from healthy samples. The amplified product was gel purified (Au-Prep Sigma gel extraction kit) and cloned in pGEM-T easy vector system (Promega, USA). The three positive clones were sequenced (Genei Pvt. Ltd, Bangalore, India) and the data were evaluated for a consensus sequence and submitted to the GenBank database (Accession EU600215).
|
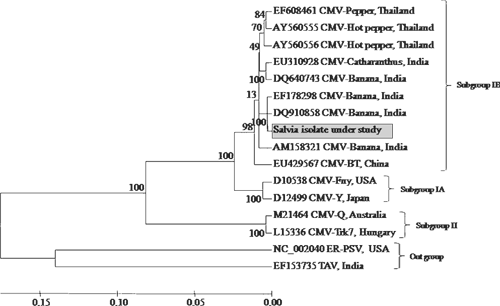
Basic Local Alignment Search Tool (BLAST) analysis of the CP gene of CMV S. splendens isolate revealed the maximum (97%) nucleotide identities with CMV isolates of subgroup IB (accessions EF178298 and DQ910858), compared with medium identities (89–88%) with isolates of CMV subgroup IA (accessions D10538 and D12499) or minimal identities (70–67%) with subgroup II (accessions M21464 and L15336) (Table 1) CMV sequences. Multiple nucleotide and deduced amino acid alignments with the available sequence were carried out using CLUSTAL-W program v1.82 (Thompson et al. 1994) and the aligned files were bootstrapped 100 times generating a neighbour-joining phylogenetic tree using Tree-Explorer. The CMV isolate under study also showed close phylogenetic relationships with Indian strains of CMV of subgroup IB, whereas more distant relationships were observed for subgroup IA and II, when analysed by MEGA 4 (Tamura et al. 2007). These cumulative studies at the biological, serological and molecular level confirm the Salvia isolate of CMV to be member of subgroup IB (Fig. 2).
|
CMV was first recorded on S. splendens from Venezuela (Debrot et al. 1974). Afterwards, CMV was also detected in China on the same species by Jian et al. (2000). To the best of our knowledge, this is the first record of CMV on S. splendens in India.
Acknowledgements
The author, A. A. Khan is thankful to the Chairman, Department of Botany for providing necessary facilities and Department of Science and Technology, New Delhi for providing financial assistance under SERC Fast Track Young Scientist Scheme.
Debrot EA,
Acosta JM, De Uzcategui RC
(1974) Natural infection of Salvia splendens with Cucumber mosaic virus. Phytopathologische Zeitschrift 80, 193–198.
| Crossref | GoogleScholarGoogle Scholar |

Jian H,
Ji-Shuang C, De-bao L
(2000) Ultrastructural alteration of host plants infected with different Cucumber mosaic virus isolates. Virologica Sinica 15, 66–72.

Srivastava A,
Chandra G, Raj SK
(2004) Coat protein and movement protein gene based molecular characterization of Amaranthus strain of Cucumber mosaic virus. Acta Virologica. English Ed. 48, 229–239.

Tamura K,
Dudley J,
Nei M, Kumar S
(2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Thompson JD,
Higgins DG, Gibson TJ
(1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Verma N,
Singh L,
Singh AK,
Kulshreshtha S,
Raikhy G,
Hallan V,
Ram R, Zaidi AA
(2005) Ornithogalum: a new host of Cucumber mosaic virus (CMV) from India. Plant Pathology 54, 256.
| Crossref | GoogleScholarGoogle Scholar |




