First record of Meloidogyne fallax in Victoria, Australia
L. Nambiar A D , M. Quader A , J. M. Nobbs B , J. A. Cobon C , P. R. Campbell C and L. M. Gulino CA Department of Primary Industries Victoria, 621 Burwood Highway, Knoxfield, Vic. 3180, Australia.
B South Australian Research and Development Institute, Field Crops Nematology Unit, LMB 100, Glen Osmond, SA 5064, Australia.
C Department of Primary Industries and Fisheries, 80 Meiers Road, Indooroopilly, Qld 4068, Australia.
D Corresponding author. Email: lila.nambiar@dpi.vic.gov.au
Australasian Plant Disease Notes 3(1) 141-142 https://doi.org/10.1071/DN08054
Submitted: 22 July 2008 Accepted: 29 September 2008 Published: 9 October 2008
Abstract
Meloidogyne fallax was detected for the first time in Victoria, Australia. It was found on potato tubers in the Cora Lynn and Toolangi districts of Victoria. The identification was based on morphological observations and molecular tests.
Nematodes of the genus Meloidogyne that cause root-knots are considered to be the world’s most damaging nematodes. Two major pest species, M. fallax and M. chitwoodi (Golden et al. 1980) are closely related and difficult to distinguish morphologically. Of these two, M. fallax is found in South Australia (Nobbs et al. 2001) and Western Australia (Vanstone and Nobbs 2007). It is proposed that M. fallax presents a photosanitary risk similar to that of the exotic M. chitwoodi (EPPO/CABI 1997). It is therefore necessary to identify these nematodes accurately based on both morphology and molecular tests.
In March 2006 diseased potato tubers from the Cora Lynn and Toolangi districts of Victoria, Australia were submitted to the Crop Health Services Unit of the Department of Primary Industry Victoria, Knoxfield for diagnosis. The tubers showed symptoms of external galling and internal necrosis just below the skin. The galls were dissected and teased out in water. A large number of eggs, juveniles, female and male nematodes were recovered. Female nematodes were used to prepare permanent slides for perineal patterns while juveniles and male nematodes were fixed in formalin–acetic fixative (4 : 1) (Hooper 1970) and transferred to anhydrous glycerol by the glycerol–ethanol method (Seinhorst 1959) for permanent microscope slide mounts.
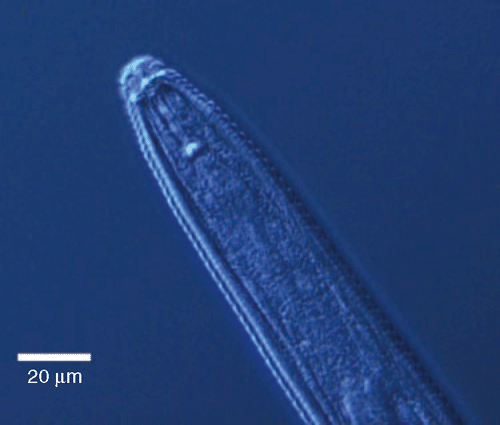
The male nematode head region was offset from body and marked with two distinct annules (Fig. 1), which is similar to that of M. chitwoodi. Measurements of juvenile stylet and total body length matched the original description of M. fallax. Perineal patterns observed were similar to that of M. hapla except that there were no punctations present in the anal region (Fig. 2) but matched the descriptions of M. fallax and M. chitwoodi. The preserved slides have been deposited in the Victorian Plant Pathology Herbarium, VPRI 33973, 33974, 33975, 33976, 33977 and 33978. Some of the observations were similar for both M. chitwoodi and M. fallax species and both of these species are known to attack potatoes. Therefore, molecular identification of specimens was also carried out to clarify the morphological identification.
|

|
The DNA was extracted from eight females selected randomly from two potato tubers using a phenol : chloroform method (Sambrook et al. 1989). Initially, published primers (Zijlstra 1997) were used to amplify a diagnostic PCR product from genomic DNA of M. fallax but despite several attempts no PCR product was found. Therefore, two primers were designed from sequences of M. fallax in GenBank (accession number GI33694309) using the computer program ‘Primer3’ (http://frodo.wi.mit.edu/, verified 30 September 2008). These primers are 18S-3-fallax (5′TTCGAGAAATTTGGGGACTG3′) and 28S-fallax (5′CGGGTGATCTCGACTGTT3′). Both primers showed specificity only to the sequences of M. fallax in the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/, verified 30 September 2008) sequence database.
The final volume of 25 µL PCR reaction contained 18.05 µL water, 2.5 µL 10 × PCR buffer, 0.75 µL 50 mM MgCl2, 0.5 µL 10 mM dNTP mix, 1 µL of 10 µM each primer, 0.5 µL Platinum® Taq (5 U/µL) DNA polymerase and 1 µL template DNA (12–19 ng/µL). A reaction without template DNA was run as negative control.
The PCR reaction was as follows: an initial temperature at 95°C for 2 min then 40 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s. A final extension period for 2 min was performed at the end of 40 cycles. The PCR products were run on a 1.5% agarose gel for 1 h and then visualised under UV light after ethidium bromide staining.
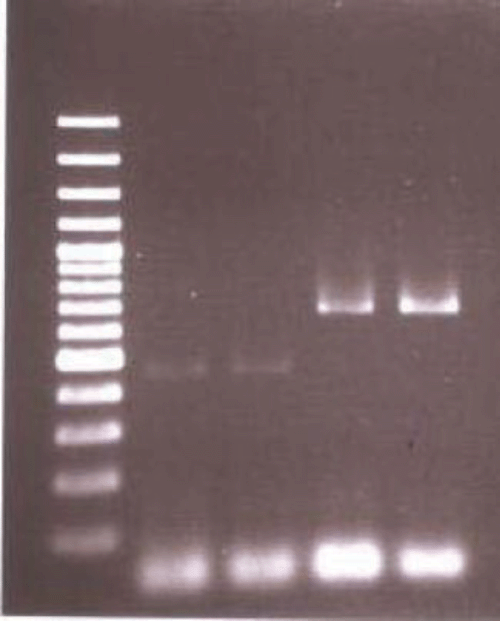
The new primers produced a 647-bp band across all DNA from the eight females of M. fallax. These PCR products were purified with a DNA Cleanup Kit (Qiagen) and sequenced (using the commercial facility of Flinders University, South Australia) for further confirmation. BLAST (NCBI) search for these sequences showed 100% similarity among themselves and 99.69% similarity with sequences of M. fallax in GenBank. These sequences have been submitted to GenBank (accession numbers EU252015–EU252022).
A single step multiplex PCR using the four diagnostic primers developed by Wishart et al. (2002) for M. fallax and M. chitwoodi and, following their method, was also used to amplify DNA from several adult females to identify species by length polymorphism. A PCR product of ~700 bp was obtained using species specific IGS primers. The size of PCR product was consistent with M. fallax (670 bp) and not M. hapla (440 bp) nor M. chitwoodi (540 bp) (Fig. 3). PCR products were gel purified using the QIAEX ll Gel Extraction Kit (Qiagen) and cloned into a pCR-TOPO vector (Invitrogen), transformed by electroporation into TOP 10 E. coli cells (Invitrogen) as per manufacturer’s instructions. Plasmid DNA was purified using QIAprep Spin Miniprep kit (Qiagen), and sequenced using ABI Big Dye V3.1. Of eight independently sequenced clones, six were identical to the M. fallax sequence on GenBank (accession number AJ421703) and two were 98.6% similar. This sequence variation is most probably due to normal population variability of the highly variable IGS region, although this is difficult to determine, as these sequences are only the second published record of this gene region in M. fallax. This sequence also has been submitted to GenBank (accession number EU711351).
|
Based on our morphological and molecular species identification, it can be concluded that the root-knot nematodes detected on potato tubers were M. fallax. This is the first record of M. fallax in Victoria.
Acknowledgements
The authors would like to thank Dr Caroline Donald and Tony Slater from DPI Victoria for providing the infected potato tubers for nematode species identification. Our sincere gratitude to Dr Farhat Shah of Crop & Food Research Ltd, New Zealand for providing M. fallax DNA.
Golden AM,
O’Bannon JH,
Santo GS, Finley AM
(1980) Description and SEM observations of Meloidogyne chitwoodi n. sp. (Meloidogynidae). A root-knot nematode on potato in the Pacific Northwest. Journal of Nematology 12, 319–327.
[Verified 30 September 2008]
Wishart J,
Phillips MS, Blok VC
(2002) Ribosomal intergenic spacer: A polymerase chain reaction diagnostic for Meloidogyne chitwoodi, M. fallax and M. hapla. Phytopathology 92(8), 884–892.
| Crossref | GoogleScholarGoogle Scholar |

Zijlstra C
(1997) A first PCR assay to identify Meloidogyne hapla, M. chitwoodi and M. fallax, and to sensitively differentiate them from each other and from M. incognita in mixtures. Fundamental and Applied Nematology 20, 505–511.



