First report of a begomovirus infecting the ornamental plant Vinca minor L.
M. Saleem Haider A D E , M. Tahir A , A. Saeed A , S. Ahmed B , R. Parveen C and N. Rashid AA School of Biological Sciences, University of the Punjab, Quaid-i-Azam Campus, Lahore, Pakistan.
B Institute of Mycology and Plant Pathology, University of the Punjab, Lahore, Pakistan.
C University College of Agriculture, Bahauddin Zakaryia University, Multan, Pakistan.
D Current address: Deptartment of Cell and Systems Biology, University of Toronto, Ontario, Canada.
E Corresponding author. Email: saleem.haider@utoronto.ca
Australasian Plant Disease Notes 3(1) 150-151 https://doi.org/10.1071/DN08058
Submitted: 1 May 2008 Accepted: 8 November 2008 Published: 20 November 2008
Abstract
Vinca minor L., an ornamental plant with symptoms typical of a begomovirus was found in the vicinity of the School of Biological Sciences, Lahore, Pakistan. Symptomatic and apparently healthy leaf samples were collected and subjected to DNA extraction and polymerase chain reaction, using universal primers conserved for the coat protein region of begomoviruses. An expected size product of ~0.78 kb was amplified from symptomatic leaf samples while healthy plant samples gave no amplification. The product was cloned and sequenced in its entirety. Basic Local Alignment Search Tool analysis showed 93% nucleotide sequence identity with Pedilanthus leaf curl virus originating from Pakistan. Our results indicate that this is the first report of a begomovirus infecting V. minor in Pakistan.
Vinca minor L. (common name Periwinkle, Sadabahar; family Apocynaceae) is a vine sub-shrub plant commonly grown as an ornamental plant in orchards and house lawns in Pakistan. During a survey for begomoviruses in ornamental plants, symptoms typical of begomoviruses, including yellowing, leaf curl and distorted leaves, were observed on the V. minor plants (Fig. 1). The infection was estimated to affect 30–40% of plants surveyed, based on visual observation of symptoms.

|
Total DNA from two symptomatic and one healthy leaf sample of V. minor was extracted by the cetyl trimethyl ammonium bromide (CTAB) method (Doyle and Doyle 1990). The presence of a begomovirus was confirmed by polymerase chain reaction (PCR) amplification using a degenerate primer pair, CPF 5′-ATG(C/A/T)(G/C)(G/C/A)AAGCG(A/T)(C/A)C(A/C)G(G/C)(A/C)GATAT-3′ and CPR 5′-TTAATT(T/G/C)(C/G/A)(A/T/C)(A/T/G)A(C/T)(A/T/C)(G/C)(C/A/T)(A/G)TCATA(G/A)AA(A/G)TA-3′, designed from published sequences of begomoviruses to conserved regions of the coat protein genes from the Old World (Haider et al. 2006). An expected size product of ~0.78 kb was amplified from symptomatic samples but not from the healthy plant sample. The product was cloned from one of the samples into a T/A cloning vector, pTZ57R/T (Fermentas Inc.), according to the manufacturer’s instructions and sequenced in its entirety with no ambiguity remaining. Sequences were determined by dideoxynucleotide chain-termination sequencing using GenomeLab Dye Terminator Cycle Sequencing kits (Beckman Coulter) on a Beckman Coulter automated sequencer (CEQ 8000). Sequences were assembled and analysed using DNASTAR (Lasergene). Multiple sequence alignments and a phylogenetic tree were produced using ClustalX. A phylogenetic tree was manipulated and printed using Treeview (Page 1996) and submitted to the EMBL database under the accession number AM292303. Comparison of the sequence isolated from V. minor with sequences available in the databases showed a maximum of 93% nucleotide sequence identity with a previously described begomovirus from Pakistan, Pedilanthus leaf curl virus-Pedilanthus (Pakistan:2004, abbreviated as PedLCV-Ped [PK:Mul:04], accession number AM712436; Tahir et al. 2008), followed by 92% nucleotide sequence identity with PedLCV-Tomato ([PK:RYK:Tom:04], accession number DQ116884).
According to recent descriptions on demarcation and nomenclature of Geminivirus strains, Fauquet et al. (2008) have outlined viruses with nucleotide identities <93% across the whole genome as strains and >94% as variants of that strain in that species, although biological data such as host range and symptom expression can also be used to define a strain. Since the virus isolate from V. minor showed highest identity (93%) with PedLCV-Ped [PK:Mul:04] across 0.78 kb of the coat protein gene, it is likely to be a strain of PedLCV-Ped [PK:Mul:04]. Conclusive nomenclature of the begomovirus isolated from V. minor will be based on its complete genome nucleotide sequence.
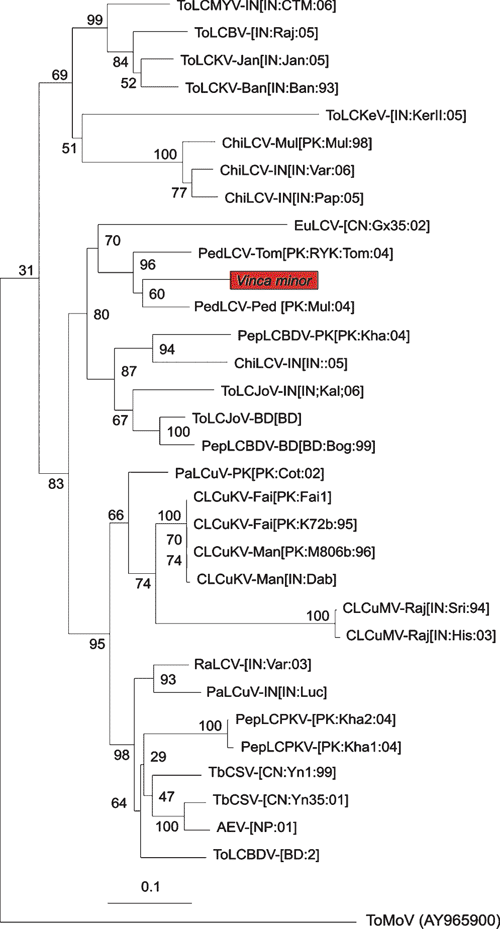
A phylogenetic analysis of the complete coat protein nucleotide sequence isolated from V. minor with the most similar sequences in the databases (determined by Basic Local Alignment Search Tool analysis) is shown in Fig. 2. This confirms close similarity with PedLCV. This is the first report of V. minor being infected with a begomovirus. An evergreen plant infected with a begomovirus is a serious threat to other economically important crops as it acts as a permanent reservoir of virus. Future studies will be focussed on the amplification of the complete genome of the virus, its phylogeny, and a detailed survey of its distribution and host range in distinct locations of the country.
|
Doyle JJ, Doyle JL
(1990) Isolation of plant DNA from fresh tissues. Focus 12, 13–15.

Fauquet CM,
Briddon RW,
Brown JK,
Moriones E,
Stanley J,
Zerbini M, Zhou X
(2008) Geminivirus strain demarcation and nomenclature. Archives of Virology 153, 783–821.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Haider MS,
Tahir M,
Latif S, Briddon RW
(2006) First report of Tomato leaf curl New Delhi virus infecting Eclipta prostrata in Pakistan. Plant Pathology 55, 285.
| Crossref | GoogleScholarGoogle Scholar |

Page RDM
(1996) TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12, 357–358.
|
CAS |
PubMed |

Tahir M,
Haider MS,
Iqbal J, Briddon RW
(2008) Association of a distinct begomovirus and a betasatellite with leaf curl symptoms of Pedilanthus tithymaloides. Journal of Phytopathology
,



