First report of ‘Candidatus Phytoplasma asteris’ (16SrI) associated with little leaf of cotton and luffa in India
S. Kumar A B , V. Singh A and S. Lakhanpaul AA Department of Botany, University of Delhi, Delhi-110007, India.
B Corresponding author. Email: sach_inom@yahoo.co.in
Australasian Plant Disease Notes 5(1) 117-119 https://doi.org/10.1071/DN10043
Submitted: 28 June 2010 Accepted: 6 October 2010 Published: 17 November 2010
Abstract
Cotton and luffa plants with little leaf symptoms were observed during February 2010 at New Delhi, India. Nested polymerase chain reaction with phytoplasma 16S rDNA-specific primers efficiently detected the pathogen in the diseased plants. Sequence comparisons showed homology with the members of ‘Candidatus Phytoplasma asteris’, group 16SrI. Phylogenetic analysis also assigned the cotton and luffa little leaf phytoplasma to the 16SrI cluster.
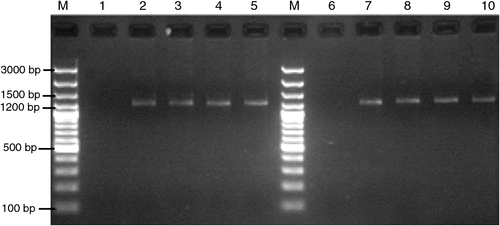
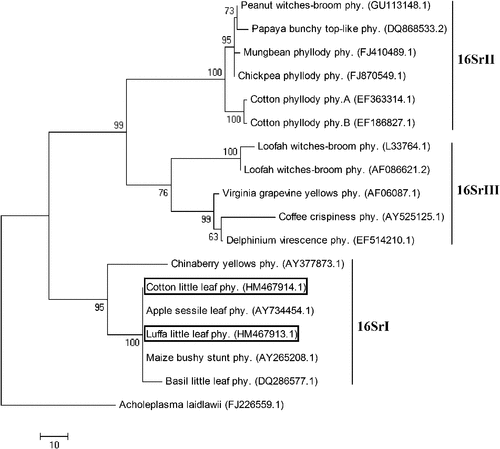
The Malvaceae member Gossypium hirsutum, commonly known as upland cotton is a world-wide valuable fibre-yielding crop. Luffa cylindrica belongs to the family Cucurbitaceae and is widely cultivated as a young edible vegetable crop and a fibre crop from mature fruits. During February 2010, these two plant species were observed to have little leaf symptoms at New Delhi, India. Leaves showed reduction in size to nearly one-third in cotton while to nearly one-fifth in luffa (Fig. 1). Such symptoms have previously been linked to phytoplasma diseases. To identify the presence of the pathogen, petioles from four symptomatic and two symptomless plants were collected for DNA isolation using cetyltrymethyl ammonium bromide as previously described (Saghai-Maroof et al. 1984). These were used as templates in a nested polymerase chain reaction (PCR) using phytoplasma-specific 16S rDNA primers P1/P7 (Deng and Hiruki 1991). The PCR products from the first round were diluted (1 : 30) and used in the subsequent round with R16F2/R2 (Gundersen and Lee 1996). Amplicons of expected size (~1.25 Kb) were observed in all the samples from symptomatic plants, while they were absent in reactions with the symptomless samples (Fig. 2). The PCR product from each sample was gel extracted, purified (QIAquick gel extraction kit, Qiagen, Germantown, MD, USA) and directly sequenced. Analysis of both the sequences using the BLAST tool from http://www.ncbi.nlm.nih.gov/ showed 99% sequence similarity to several 16SrI phytoplasma members, namely sesame phyllody (AB558132.1), Japanese spurge yellows (AB551736.1), Eryngium foetidum witches’ broom (GU113155.1), periwinkle little leaf (GU113147.1), mulberry yellow dwarf (GQ249410.1) phytoplasmas. The partial 16S rDNA sequences were submitted to GenBank with Accession Nos. HM467914 and HM467913 for cotton and luffa little leaf phytoplasma, respectively. A dendrogram was constructed using maximum parsimony method of MEGA software v.4.1 (Tamura et al. 2007) with phytoplasma 16S rDNA sequences as raw data. The evolutionary tree depicted the cotton and luffa little leaf phytoplasma to cluster within the 16SrI clade (Fig. 3), inferring their close relationship with the aster yellow members. The earlier reported cotton (EF186827.1, EF363314.1) and luffa (AF086621.2, L33764.1) phytoplasmas took distinct positions in the 16SrII and 16SrIII cluster. Thus, the sequence and phylogenetic analysis assign the phytoplasma associated with little leaf of cotton and luffa to ‘Ca. Phytoplasma asteris’.

|
|
|
Molecular investigations to date have shown phytoplasmas to be associated with cotton phyllody in Italy, Burkina Faso, Mali (Khan et al. 2002; Martini et al. 2007; Marzachi et al. 2009) and luffa witches’ broom in Brazil (Montano et al. 2007). This is the first record of a phytoplasma associated with both these economically important crops in the Indian subcontinent. Additionally, cotton and luffa are hereby reported as the two new hosts of 16SrI group phytoplasmas.
References
Deng S, Hiruki C (1991) Amplification of 16S rRNA genes from culturable and non-culturable mollicutes. Journal of Microbiological Methods 14, 53–61.| Amplification of 16S rRNA genes from culturable and non-culturable mollicutes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38XhsVCqt74%3D&md5=1310ad025a0ab382b665ffd8a498fdfeCAS |
Gundersen DE, Lee I-M (1996) Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathologia Mediterranea 35, 144–151.
Khan A, Botti S, Al-Subhi A, Gundersen-Rindal D, Bertaccini A (2002) Molecular identification of a new phytoplasma associated with alfalfa witches’-broom in Oman. Phytopathology 92, 1038–1047.
| Molecular identification of a new phytoplasma associated with alfalfa witches’-broom in Oman.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XotlGitr0%3D&md5=de0da0932bb5d9b9b23a9ceb2c25c32dCAS | 18944213PubMed |
Martini M, Lee I-M, Bottner KD, Zhao Y, Botti S, Bertaccini A, Harrison NA, Carraro L, Marcone C, Khan AJ, Osler R (2007) Ribosomal protein gene-based phylogeny for finer differentiation and classification of phytoplasmas. International Journal of Systematic and Evolutionary Microbiology 57, 2037–2051.
| Ribosomal protein gene-based phylogeny for finer differentiation and classification of phytoplasmas.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXht1Wms7rO&md5=1405393a3d210ab281892e78c92dfd7aCAS | 17766869PubMed |
Marzachi C, Coulibaly A, Coulibaly N, Sangare A, Diarra M, Gregorio TD, Bosco D (2009) Cotton virescence phytopasma and its weed reservoir in Mali. Journal of Plant Pathology 91, 717–721.
Montano HG, Brioso PST, Cunha Jn JO, Figueiredo DV, Pimentel JP (2007) First report of group 16SrIII phytoplasma in loofah (Luffa cylindrica). Bulletin of Insectology 60, 277–278.
Saghai-Maroof MA, Soliman KM, Jorgenson RA, Allard AW (1984) Ribosomal spacer-length polymorphism in barley: Mendelian inheritance, chromosomal creation and population dynamics. Proceedings of the National Academy of Sciences of the United States of America 81, 8014–8018.
| Ribosomal spacer-length polymorphism in barley: Mendelian inheritance, chromosomal creation and population dynamics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2MXptlWitg%3D%3D&md5=ce69c321e11d0c84e6fad3c342235839CAS | 6096873PubMed |
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599.
| MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXpsVGrsL8%3D&md5=d8ad169b9ccd0e7d548d166c6370749dCAS | 17488738PubMed |


