Henipaviruses: bat-borne paramyxoviruses
Sarah Edwards A and Glenn A Marsh A BA Australian Animal Health Laboratory
CSIRO Health and Biosecurity
5 Portarlington Road
East Geelong, Vic. 3219, Australia
Tel: +61 3 5227 5125
Fax: +61 3 5227 5555
B Email: glenn.marsh@csiro.au
Microbiology Australia 38(1) 4-7 https://doi.org/10.1071/MA17003
Published: 21 February 2017
Found on every continent except Antarctica, bats are one of the most abundant, diverse and geographically widespread vertebrates globally, making up approximately 20% of all known extant mammal species1,2. Noted for being the only mammal with the ability of powered flight, bats constitute the order Chiroptera (from the Ancient Greek meaning ‘hand wing’), which is further divided into two suborders: Megachiroptera known as megabats or flying foxes, and Microchiroptera comprising of echolocating microbats1,3.
Known for their important role in the pollination of flora, seed dispersal and insect population control, bats also play a key role in the spread and perseverance of many notable zoonotic viruses which cause severe disease and potentially fatal outcomes for humans, livestock and many other species1,4. In particular, megabats belonging to the genus Pteropus, are important natural reservoirs for many significant pathogenic viruses such as Hendra virus (HeV), Nipah virus (NiV), SARS Coronavirus, Australian bat Lyssavirus and Menangle virus4–9.
The emergence of Henipaviruses
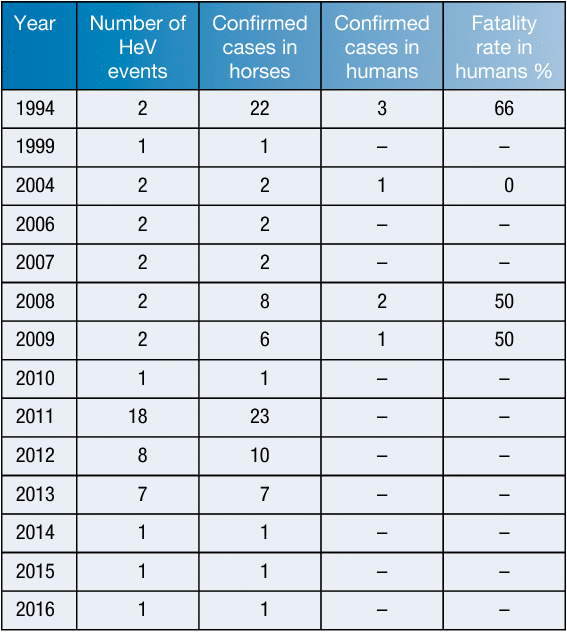
HeV was first observed in 1994 during a severe respiratory disease outbreak in Brisbane, Australia, in which 13 of 21 infected horses died within a two week period10. Additionally, two humans who frequently had extensive contact with the affected horses were confirmed as infected, with one later dying as a result10,11. Since the emergence of HeV, sporadic outbreaks have occurred within the equine population of Australia, which have resulted in four human fatalities, as well as the death or euthanasia of over 84 horses and two dogs12,13 (Table 1). In late 2012 an HeV vaccine, based on the G glycoprotein, was released for use in horses14. Since this time, all identified cases of HeV have been in unvaccinated horses.
|
Following initial HeV outbreaks, the closely related NiV emerged in 1998 amongst the pig and human population in Peninsular Malaysia. During the outbreak, 105 people died from 265 cases of infection with febrile illness and encephalitis15. The disease was also later recorded in Singapore, transmitted via the importation of infected swine from the outbreak region of Malaysia. Over 1 million pigs were culled in a large-scale effort to eradicate the causative pathogen – an effort that has proven successful in this region11,16. Three years later, disease with hallmarks of febrile neurological symptoms resulted in the death of 9 people in a Bangladeshi village. An investigation into subsequent outbreaks showed a reaction of antibodies to NiV antigens17, however, unlike the Malaysian NiV outbreak, pigs were not the intermediate and amplifying host. Instead, human-to-human and bat-to-human transmission routes were involved in NiV outbreaks occurring within the Bangladesh region – events not previously observed of the henipaviruses11. Furthermore, the more recent Nipah outbreaks demonstrated a notably higher fatality ratio of 74%, compared to 38.5% for the Malaysia outbreak15,18.
During 2014, severe illness among humans and horses in southern Philippines was attributed to henipavirus infections19. Seventeen cases met the case definition with 2 survivors, of these seven had participated in horse slaughtering and horse meat consumption. Horse-to-human and human-to-human transmission occurred.
Additionally, although recovery from HeV and NiV infection is possible, relapsed encephalitis has been shown to occur in 3–7% of individuals anywhere from months to years following recovery from acute infection20.
Bats as hosts to the Henipaviruses
Following the 1994 Brisbane HeV outbreak, a large serological survey of horses showed no evidence of HeV infection outside the index property21, suggesting horses were not commonly infected with these viruses. An extensive sampling exercise of native and introduced animal species was carried out with antibodies to HeV being detected in all four mainland Australian flying fox species: the Black flying fox (Pteropus alecto), Grey-headed flying fox (P. poliocephalus), Spectacled flying fox (P. conspicilla- tus) and the Little red flying fox (P. scapulatus)9,22. Further research has resulted in isolation of HeV directly from pteropid bats23,24.
Following the 1998 Malaysian NiV outbreak, investigations of bat colonies across a large area of Peninsular Malaysia revealed evidence of NiV antibodies in the two pteropid species found in Malaysia: the Island flying fox (P. hypomelanus) and the Malayan flying fox (P. vampyrus)25,26. NiV was also isolated from urine and partially eaten fruit collected from a colony of P. hypomelanus on Tioman Island, off the coast of Peninsular Malaysia27.
Following the emergence of NiV in India and Bangladesh in 2001, P. giganteus, a flying fox found across the Indian subcontinent, was shown to have antibodies to NiV17. To date, no NiV isolate has been reported from bats in either Bangladesh or India.
Although disease associated with HeV and NiV has only been observed in Australia and South and South-east Asia, serological and molecular evidence of Henipavirus have been reported from many different countries, including Africa28–31 and Asia32. This includes 2 full genome sequences that have been obtained, one from a bat in Ghana (Kumasi virus)33 and the other from a Chinese rat (Mòjiāng virus)34.
There is evidence that less pathogenic Henipaviruses may also be circulating in Australia, exemplified by the newly identified CedPV virus35. This was observed to share a similar genome size and organisation with HeV and NiV, showed cross-reactivity with henipavirus antigens, and also used the same host cell receptor for infection. Despite these similarities, disease was not observed with CedPV infection in various animal models in which a lethal disease results from HeV or NiV challenge. This is thought to be due to differences in its phosphoprotein gene in which it lacks RNA editing sequences and a highly conserved V open reading frame35, whose protein product is responsible for modulating the host innate immune response32.
The molecular biology of Henipaviruses
HeV, NiV and CedPV belong to the genus Henipavirus (order Mononegavirales, family Paramyxoviridae). HeV and NiV were the first observed zoonotic Paramyxoviruses resulting from spillover events of Pteropid bats36. High virulence, human susceptibility, broad host species range, and a lack of human vaccines and therapeutic treatment has resulted in HeV and NiV being restricted to biosafety level four (BSL4) containment – the only Paramyxoviruses to be restricted to this level35,37,38. The lack of a human vaccine and licensed therapeutics necessitates on-going research to understand these viruses and the virulence determinants.
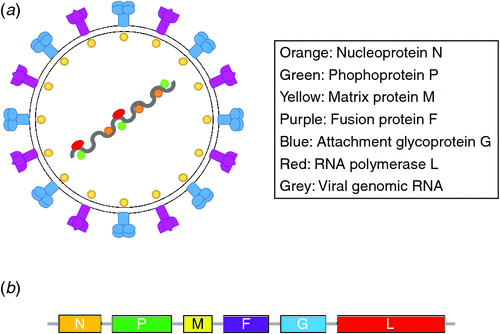
Members of the Paramyxoviridae family are enveloped, non-segmented negative-stranded RNA viruses which can cause a range of respiratory and systemic disease in both humans and animals. Paramyxovirus genomes share similar ultrastructural appearance, and organisation of genes contained within a genome which can range from 15–20 kilobases (kb), with henipaviruses having a genome size of 18.2 kb39,40. Henipavirus genomes encode six major proteins. Reading in order from 3’ to 5’ on the antigenome they are: nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), attachment glycoprotein (G), and RNA-dependent RNA polymerase (L)39,41 (Figure 1).
|
Essential for the replication and transcription of paramyxoviruses, the genomic RNA is encapsidated by the N protein, the most abundant protein in the purified virion. The N protein associates with both the P and L proteins, forming the ribonucleoprotein (RNP) complex, and it is this complex that serves as a template for replication42,43. Following transcription and translation of viral proteins, the M protein mediates the association of the plasma membrane with the ribonucleoprotein and glycoproteins F and G. After assembly, the newly matured virion is then ready to bud and be released from the host cell44.
The attachment glycoprotein (G glycoprotein/HN haemaglutinin-neuraminidase/H haemaglutinin; depending on the virus), is essential for the initiation of infection by recognising and attaching to membrane-bound host cell receptors. In the case of the henipaviruses, the G glycoprotein attaches to receptors ephrin B2 and B345–47. The binding of the G protein to the ephrin host cell receptor facilitates the F protein-dependent fusion event, however the exact signalling cascade from the bound G glycoprotein to the fusion-competent F protein is not well understood47. Due to the enveloped nature of the paramyxoviruses, it is essential that fusion-competent F proteins generate membrane fusion between the virus envelope and the plasma membrane of the receptive host cell through a pathway of conformational changes in the viral envelope proteins38.
Similar to other Paramyxoviridae genomes, the henipavirus genomes strictly follow the ‘rule of six’ which requires that the genome has a nucleotide length of a multiple of six (also known as a polyhexameric length). The importance of nucleotide numbers being a multiple of six is thought to be due to the nucleocapsid proteins interacting with precisely six nucleotides therefore forming a hexamer within the viral RNP complex48,49.
Given the global zoonotic threat posed by these viruses, there is a great importance to further understand these viruses. However, given that these agents are amongst the most dangerous pathogens known, research methods have been developed to allow many of these agents to be handled outside of BSL4 containment. Reverse genetics has advanced the study of negative-sense RNA viruses, and is a commonly used tool for the in vitro study of negative-stranded RNA virus replication by allowing the manipulation of genes within the viral genome50,51. These systems rely on the transfection of ‘helper’ plasmids which each encode a gene required for viral replication and gene expression. The genes encoded on these expression vectors are present as cDNA, and are therefore not infectious50,52,53.
Future perspectives
The infection of HeV and NiV in horses and pigs, respectively, demonstrate the potential threat of dangerous bat-borne zoonotic agents to livestock as the intermediate and amplifying host for transmission to humans. Factors contributing to the emergence of these viruses into the human population include deforestation, closer living proximities between humans and bats, and a greater yield demand and turnover of livestock farming practices. Considering the close human interaction with livestock within current farming practices globally, livestock interaction with the bat population is of high importance for both intervention and countermeasure strategies to prevent incidences of human infection. In addition to surveillance of both bat populations and any circulating zoonotic agents they may harbour, further study of these viruses on a molecular biology and immunology scale is needed. Broadening our understanding of key factors such as bat immunity, virus transmission, host response and pathogenesis, could make a significant contribution to the development of vaccines and therapeutic treatments for these highly pathogenic bat-borne infections.
References
[1] Calisher, C.H. et al. (2006) Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19, 531–545.| Bats: important reservoir hosts of emerging viruses.Crossref | GoogleScholarGoogle Scholar |
[2] Teeling, E.C. et al. (2005) A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307, 580–584.
| A molecular phylogeny for bats illuminates biogeography and the fossil record.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmslOitA%3D%3D&md5=0f95f418e074df572a69381abe93f681CAS |
[3] Holland, R.A. et al. (2004) Echolocation signal structure in the Megachiropteran bat Rousettus aegyptiacus Geoffroy 1810. J. Exp. Biol. 207, 4361–4369.
| Echolocation signal structure in the Megachiropteran bat Rousettus aegyptiacus Geoffroy 1810.Crossref | GoogleScholarGoogle Scholar |
[4] Edson, D. et al. (2015) Flying-fox roost disturbance and Hendra virus spillover risk. PLoS One 10, e0125881.
| Flying-fox roost disturbance and Hendra virus spillover risk.Crossref | GoogleScholarGoogle Scholar |
[5] Smith, I. and Wang, L.F. (2013) Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 3, 84–91.
| Bats and their virome: an important source of emerging viruses capable of infecting humans.Crossref | GoogleScholarGoogle Scholar |
[6] Poon, L.L. et al. (2005) Identification of a novel coronavirus in bats. J. Virol. 79, 2001–2009.
| Identification of a novel coronavirus in bats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXitlejtrc%3D&md5=ba7696f03e3b6228a95a9ed15f515b3bCAS |
[7] Fraser, G.C. et al. (1996) Encephalitis caused by a Lyssavirus in fruit bats in Australia. Emerg. Infect. Dis. 2, 327–331.
| Encephalitis caused by a Lyssavirus in fruit bats in Australia.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2s7isF2gsA%3D%3D&md5=3424d46c23be0a34426ebbb25f14d047CAS |
[8] Chant, K. et al. (1998) Probable human infection with a newly described virus in the family Paramyxoviridae. The NSW Expert Group. Emerg. Infect. Dis. 4, 273–275.
| Probable human infection with a newly described virus in the family Paramyxoviridae. The NSW Expert Group.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK1c3osVaiug%3D%3D&md5=8e85c4ff40f4575e22346607eaa38670CAS |
[9] Young, P. (1996) Possible reservoir host of equine morbillivirus identified. Commun. Dis. Intell. 20, 262.
[10] Murray, K. et al. (1995) A morbillivirus that caused fatal disease in horses and humans. Science 268, 94–97.
| A morbillivirus that caused fatal disease in horses and humans.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXkvV2ksLw%3D&md5=72172f32c43751df3f4a48c889ff6688CAS |
[11] Field, H. et al. (2001) The natural history of Hendra and Nipah viruses. Microbes Infect. 3, 307–314.
| The natural history of Hendra and Nipah viruses.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3M3jvVaqtA%3D%3D&md5=e3aea7888e60b0ac1708049bff5e544aCAS |
[12] Mendez, D. et al. (2013) Response of Australian veterinarians to the announcement of a Hendra virus vaccine becoming available. Aust. Vet. J. 91, 328–331.
| Response of Australian veterinarians to the announcement of a Hendra virus vaccine becoming available.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3sfkt1eksw%3D%3D&md5=a97e57bda00a8e86c309fdd658b2bfecCAS |
[13] Croser, E.L. and Marsh, G.A. (2013) The changing face of the henipaviruses. Vet. Microbiol. 167, 151–158.
| The changing face of the henipaviruses.Crossref | GoogleScholarGoogle Scholar |
[14] Middleton, D. et al. (2014) Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg. Infect. Dis. 20, 372–379.
| Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health.Crossref | GoogleScholarGoogle Scholar |
[15] Chua, K.B. (2003) Nipah virus outbreak in Malaysia. J. Clin. Virol. 26, 265–275.
| Nipah virus outbreak in Malaysia.Crossref | GoogleScholarGoogle Scholar |
[16] Chua, K.B. et al. (2000) Nipah virus: a recently emergent deadly paramyxovirus. Science 288, 1432–1435.
| Nipah virus: a recently emergent deadly paramyxovirus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXjslGqsrw%3D&md5=94bb08eac69c7355bc8fc6b249ffdcc8CAS |
[17] Hsu, V.P. et al. (2004) Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 10, 2082–2087.
| Nipah virus encephalitis reemergence, Bangladesh.Crossref | GoogleScholarGoogle Scholar |
[18] Chadha, M.S. et al. (2006) Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 12, 235–240.
| Nipah virus-associated encephalitis outbreak, Siliguri, India.Crossref | GoogleScholarGoogle Scholar |
[19] Ching, P.K. et al. (2015) Outbreak of henipavirus infection, Philippines, 2014. Emerg. Infect. Dis. 21, 328–331.
| Outbreak of henipavirus infection, Philippines, 2014.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XitVKisLzK&md5=e0682e3a14b4b9fc90c8d3613fbbcca7CAS |
[20] Tan, C.T. et al. (2002) Relapsed and late-onset Nipah encephalitis. Ann. Neurol. 51, 703–708.
| Relapsed and late-onset Nipah encephalitis.Crossref | GoogleScholarGoogle Scholar |
[21] Ward, M.P. et al. (1996) Negative findings from serological studies of equine morbillivirus in the Queensland horse population. Aust. Vet. J. 74, 241–243.
| Negative findings from serological studies of equine morbillivirus in the Queensland horse population.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2s%2FkvFeisA%3D%3D&md5=6130e419c65cb55942ca3403e696eff0CAS |
[22] Young, P.L. et al. (1996) Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg. Infect. Dis. 2, 239–240.
| Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2s%2FmsVWitQ%3D%3D&md5=6cea582b58a3f0ddc9ab0529e539c2eaCAS |
[23] Halpin, K. et al. (2000) Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 81, 1927–1932.
| Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXlsFOhtLo%3D&md5=bd671cee0260ddf9fdb92b1382328605CAS |
[24] Smith, I. et al. (2011) Identifying Hendra virus diversity in pteropid bats. PLoS One 6, e25275.
| Identifying Hendra virus diversity in pteropid bats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlCjs7fO&md5=b4df5824352faf1b96d0c8974b0617dbCAS |
[25] Shirai, J. et al. (2007) Nipah Virus Survey of Flying Foxes in Malaysia. Jpn. Agric. Res. Q. 41, 69–78.
| Nipah Virus Survey of Flying Foxes in Malaysia.Crossref | GoogleScholarGoogle Scholar |
[26] Yob, J.M. et al. (2001) Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7, 439–441.
| Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3MzhtFOhsQ%3D%3D&md5=521e44b6cb29bf893c99af98f5434c5eCAS |
[27] Chua, K.B. et al. (2002) Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 4, 145–151.
| Isolation of Nipah virus from Malaysian Island flying-foxes.Crossref | GoogleScholarGoogle Scholar |
[28] Iehlé, C. et al. (2007) Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg. Infect. Dis. 13, 159–161.
| Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar.Crossref | GoogleScholarGoogle Scholar |
[29] Drexler, J.F. et al. (2009) Henipavirus RNA in African bats. PLoS One 4, e6367.
| Henipavirus RNA in African bats.Crossref | GoogleScholarGoogle Scholar |
[30] Hayman, D.T. et al. (2008) Evidence of henipavirus infection in West African fruit bats. PLoS One 3, e2739.
| Evidence of henipavirus infection in West African fruit bats.Crossref | GoogleScholarGoogle Scholar |
[31] Pernet, O. et al. (2014) Evidence for henipavirus spillover into human populations in Africa. Nat. Commun. 5, 5342.
| Evidence for henipavirus spillover into human populations in Africa.Crossref | GoogleScholarGoogle Scholar |
[32] Baker, K.S. et al. (2013) Novel, potentially zoonotic paramyxoviruses from the African straw-colored fruit bat Eidolon helvum. J. Virol. 87, 1348–1358.
| Novel, potentially zoonotic paramyxoviruses from the African straw-colored fruit bat Eidolon helvum.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXltV2isLg%3D&md5=522a850c2f817fd849e88afbf193f80eCAS |
[33] Drexler, J.F. et al. (2012) Bats host major mammalian paramyxoviruses. Nat. Commun. 3, 796.
| Bats host major mammalian paramyxoviruses.Crossref | GoogleScholarGoogle Scholar |
[34] Wu, Z. et al. (2014) Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg. Infect. Dis. 20, 1064–1066.
| Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012.Crossref | GoogleScholarGoogle Scholar |
[35] Marsh, G.A. et al. (2012) Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 8, e1002836.
| Cedar virus: a novel Henipavirus isolated from Australian bats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFKlurbK&md5=78f9f74f5117d823d9ab5e980a86cf99CAS |
[36] Mortlock, M. et al. (2015) Novel Paramyxoviruses in Bats from Sub-Saharan Africa, 2007–2012. Emerg. Infect. Dis. J. 21, 1840.
| Novel Paramyxoviruses in Bats from Sub-Saharan Africa, 2007–2012.Crossref | GoogleScholarGoogle Scholar |
[37] Craft, W.W. and Dutch, R.E. (2005) Sequence motif upstream of the Hendra virus fusion protein cleavage site is not sufficient to promote efficient proteolytic processing. Virology 341, 130–140.
| Sequence motif upstream of the Hendra virus fusion protein cleavage site is not sufficient to promote efficient proteolytic processing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVKjs73L&md5=a89e1ecc71c9ddbe3d9dcafa7458c9d5CAS |
[38] Smith, E.C. et al. (2012) Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion. J. Biol. Chem. 287, 30035–30048.
| Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1CgtrjF&md5=415d51661eae4f7f7c15f247e62e8161CAS |
[39] Wang, L. et al. (2001) Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3, 279–287.
| Molecular biology of Hendra and Nipah viruses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXjsFShuro%3D&md5=d734a9179a72d17942d72ee33593aec8CAS |
[40] Harcourt, B.H. et al. (2001) Molecular characterization of the polymerase gene and genomic termini of Nipah virus. Virology 287, 192–201.
| Molecular characterization of the polymerase gene and genomic termini of Nipah virus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXlvFWit7w%3D&md5=6bbc354f00d1081741940b39d5bfd651CAS |
[41] Harcourt, B.H. et al. (2000) Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271, 334–349.
| Molecular characterization of Nipah virus, a newly emergent paramyxovirus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXktVKqtL8%3D&md5=4eeccdeb261e0202d84d21f5ea212d40CAS |
[42] Buchholz, U.J. et al. (1999) Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73, 251–259.
| 1:CAS:528:DyaK1cXotFSksro%3D&md5=1fbd29345deefc1a7793cb6d586a8286CAS |
[43] Hoenen, T. et al. (2011) Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. 91, 195–208.
| Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXovF2it7w%3D&md5=c1d794c7efaa70c22001ff829059260cCAS |
[44] Patch, J.R. et al. (2007) Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol. J. 4, 1.
| Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein.Crossref | GoogleScholarGoogle Scholar |
[45] Bossart, K.N. et al. (2001) Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology 290, 121–135.
| Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXot12msr0%3D&md5=2de0d3ad6d77c6029cc6c06e4362ac8fCAS |
[46] Bonaparte, M.I. et al. (2005) Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 102, 10652–10657.
| Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXntVSiu78%3D&md5=d1a93a48b3ffd83a6ead95094e77b63fCAS |
[47] Aguilar, H.C. and Iorio, R.M. (2012) Henipavirus membrane fusion and viral entry. Curr. Top. Microbiol. Immunol. 359, 79–94.
| Henipavirus membrane fusion and viral entry.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvVCmtLzM&md5=84f6579bfd7149dc89dee5007b4fa2f3CAS |
[48] Skiadopoulos, M.H. et al. (2003) The genome length of human parainfluenza virus type 2 follows the rule of six, and recombinant viruses recovered from non-polyhexameric-length antigenomic cDNAs contain a biased distribution of correcting mutations. J. Virol. 77, 270–279.
| The genome length of human parainfluenza virus type 2 follows the rule of six, and recombinant viruses recovered from non-polyhexameric-length antigenomic cDNAs contain a biased distribution of correcting mutations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXkt1am&md5=fcc2cc6939fae6d4d858cde5d28e9df9CAS |
[49] Vulliémoz, D. and Roux, L. (2001) ‘Rule of six’: how does the Sendai virus RNA polymerase keep count? J. Virol. 75, 4506–4518.
| ‘Rule of six’: how does the Sendai virus RNA polymerase keep count?Crossref | GoogleScholarGoogle Scholar |
[50] Yoneda, M. et al. (2006) Establishment of a Nipah virus rescue system. Proc. Natl. Acad. Sci. USA 103, 16508–16513.
| Establishment of a Nipah virus rescue system.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1WmsLnK&md5=49d74de4c47cb90384226080929fe8a4CAS |
[51] Kovacs, G.R. et al. (2003) Enhanced genetic rescue of negative-strand RNA viruses: use of an MVA-T7 RNA polymerase vector and DNA replication inhibitors. J. Virol. Methods 111, 29–36.
| Enhanced genetic rescue of negative-strand RNA viruses: use of an MVA-T7 RNA polymerase vector and DNA replication inhibitors.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXkvVGqtbs%3D&md5=2a2bf1e8fbf5254bb8d43743df30a3cbCAS |
[52] Brown, D.D. et al. (2005) ‘Rescue’ of mini-genomic constructs and viruses by combinations of morbillivirus N, P and L proteins. J. Gen. Virol. 86, 1077–1081.
| ‘Rescue’ of mini-genomic constructs and viruses by combinations of morbillivirus N, P and L proteins.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjtlOmtr4%3D&md5=f7f8c92ed2b724a3fbfdb9e0ef66d62eCAS |
[53] Freiberg, A. et al. (2008) Establishment and characterization of plasmid-driven minigenome rescue systems for Nipah virus: RNA polymerase I- and T7-catalyzed generation of functional paramyxoviral RNA. Virology 370, 33–44.
| Establishment and characterization of plasmid-driven minigenome rescue systems for Nipah virus: RNA polymerase I- and T7-catalyzed generation of functional paramyxoviral RNA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtlylu7%2FL&md5=5fc2f88791b511d16fba4e96aa2737dcCAS |
Biographies
Sarah Edwards is a PhD student based at the CSIRO Australian Animal Health Laboratory. Her project is investigating the molecular mechanisms of neurological invasion of the henipaviruses in a mouse model.
Glenn Marsh is a Senior Research Scientist and Team Leader for Dangerous Pathogens in CSIRO. His research interests include development of animal models for high consequence viruses and using molecular tools to identify virulence determinants.


