Bats, bacteria and their role in health and disease
Kristin MühldorferLeibniz Institute for Zoo and Wildlife Research
Department of Wildlife Diseases
Alfred-Kowalke-Str. 17, 10315 Berlin, Germany
Tel: +49 30 5168-215
Email: muehldorfer@izw-berlin.de
Microbiology Australia 38(1) 28-29 https://doi.org/10.1071/MA17009
Published: 9 February 2017
Bats are ancient and among the most diverse mammals in terms of species richness, diet and habitat preferences, characteristics that may contribute to a high diversity of infectious agents. During the past two decades, the interest in bats and their microorganisms largely increased because of their role as reservoir hosts or carriers of important pathogens. Rapid advances in microbial detection and characterisation by high-throughput sequencing technologies have led to large genetic data sets but also improved our possibilities and speed of identifying unknown infectious agents. Assessing the risk of infectious diseases in bats and their pathological manifestation, however, is still challenging because of limited access to appropriate material and field data, and continuing limitations in wildlife diagnostics and the interpretation of genetic results. As a consequence, emerging pathogens can suddenly appear with devastating effects as happened for the white nose syndrome. To date, much research on bats and infectious agents still focusses on viruses, whilst the knowledge on bacteria and their role in disease is comparatively low.
Some bacterial pathogens, such as Bartonella and Leptospira, have been more intensively studied and it seems that bats have a remarkably high diversity of bacterial species1,2. Interestingly, genetic analyses provide continuous evidence of yet-undescribed bacterial sequences and clades that might be associated with different species of bats, their habitats, ectoparasites (only for Bartonella) or geographic origins. It is very likely that they represent novel species within the families Bartonellaceae and Leptospiraceae but need to be further characterised.
Novel bacteria belonging to the family of Pasteurellaceae were isolated from three different species of vespertilionid bats from Europe3. They were classified as members of the genus Vespertiliibacter and provide some evidence for species-specific adaptations in bats. Three further isolates that also belong to this genus have been collected recently from Nathusius’s pipistrelles (Pipistrellus nathusii) confirming the presence of diverse bacterial strains in different species of insectivorous bats (family Vespertilionidae) (Figure 1).
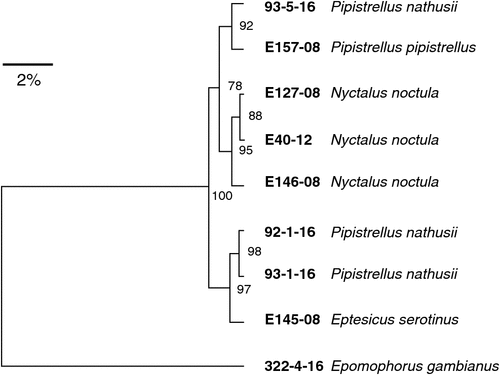
|
Bartonella, Leptospira and many Pasteurellaceae species are known pathogens that can cause serious bacterial infections in mammals. Infected bats, however, appear to be healthy or do not show specific disease symptoms suggesting that these bacteria might possibly have co-evolved with their hosts4. To date, bacterial diseases are rarely described in wild bats5. They often represent individual cases that have been caused by well-recognised bacterial pathogens such as Pasteurella multocida, Escherichia coli, Salmonella and Yersinia species. The only exception known so far is an outbreak of acute systemic pasteurellosis caused by P. multocida in a colony of wild big brown bats (Eptesicus fuscus) in Wisconsin, USA, which resulted in high mortality6. In fruit bats kept in captivity sporadic infections associated with different opportunistic bacteria (e.g. Staphylococcus aureus, Listeria or alpha-hemolytic Streptococcus spp.) (Lisa L Farina, pers. comm.) and serious outbreaks of Yersinia pseudotuberculosis7 are known to occur.
The limited number of studies and literature available on infectious diseases of bats most probably reflect challenges we face in wildlife health investigations. For example, bats roosting and foraging in close proximity to humans are more likely to be found than bats that inhabit remote areas. Predation, scavenging and the fast decomposition of bat carcasses limit the access to diseased or dead animals and strongly affect wildlife diagnostics. High-throughput sequencing has markedly enhanced our abilities to detect unknown, non-culturable infectious agents or to study host-associated microbial communities in bats8. The genetic analyses are complex and generate large amounts of sequencing data that represent numerous bacterial taxa (host microbiota, unknown bacterial species, ingested and environmental bacteria) and require judicious interpretation of results. Clearly, they provide valuable insights into the composition of the microflora and the bacterial diversity in different species of bats9,10. In perspective, basic knowledge about bats and their microorganisms will enhance our understanding of existing bacteria-host interactions and their role in health and disease.
Acknowledgement
Thanks to Nadine Jahn for her technical assistance.
References
[1] McKee, C.D. et al. (2016) Phylogenetic and geographic patterns of Bartonella host shifts among bat species. Infect. Genet. Evol. 44, 382–394.| Phylogenetic and geographic patterns of Bartonella host shifts among bat species.Crossref | GoogleScholarGoogle Scholar |
[2] Dietrich, M. et al. (2015) Leptospira and bats: story of an emerging friendship. PLoS Pathog. 11, e1005176.
| Leptospira and bats: story of an emerging friendship.Crossref | GoogleScholarGoogle Scholar |
[3] Mühldorfer, K. et al. (2014) Proposal of Vespertiliibacter pulmonis gen. nov., sp. nov. and two genomospecies as new members of the family Pasteurellaceae isolated from European bats. Int. J. Syst. Evol. Microbiol. 64, 2424–2430.
| Proposal of Vespertiliibacter pulmonis gen. nov., sp. nov. and two genomospecies as new members of the family Pasteurellaceae isolated from European bats.Crossref | GoogleScholarGoogle Scholar |
[4] Lei, B.R. and Olival, K.J. (2014) Contrasting patterns in mammal-bacteria coevolution: Bartonella and Leptospira in bats and rodents. PLoS Negl. Trop. Dis. 8, e2738.
| Contrasting patterns in mammal-bacteria coevolution: Bartonella and Leptospira in bats and rodents.Crossref | GoogleScholarGoogle Scholar |
[5] Mühldorfer, K. (2013) Bats and bacterial pathogens: a review. Zoonoses Public Health 60, 93–103.
| Bats and bacterial pathogens: a review.Crossref | GoogleScholarGoogle Scholar |
[6] Blehert, D.S. et al. (2014) Acute pasteurellosis in wild big brown bats (Eptesicus fuscus). J. Wildl. Dis. 50, 136–139.
| Acute pasteurellosis in wild big brown bats (Eptesicus fuscus).Crossref | GoogleScholarGoogle Scholar |
[7] Nakamura, S. et al. (2013) Outbreak of yersiniosis in Egyptian rousette bats (Rousettus aegyptiacus) caused by Yersinia pseudotuberculosis serotype 4b. J. Comp. Pathol. 148, 410–413.
| Outbreak of yersiniosis in Egyptian rousette bats (Rousettus aegyptiacus) caused by Yersinia pseudotuberculosis serotype 4b.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38bnt1ahuw%3D%3D&md5=28ba1061faaa34a4796b31c7fb4ee8d1CAS |
[8] Phillips, C.D. et al. (2012) Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol. Ecol. 21, 2617–2627.
| Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography.Crossref | GoogleScholarGoogle Scholar |
[9] Dietrich, M. et al. (2017) The excreted microbiota of bats: evidence of niche specialisation based on multiple body habitats. FEMS Microbiol. Lett. 364, .
| The excreted microbiota of bats: evidence of niche specialisation based on multiple body habitats.Crossref | GoogleScholarGoogle Scholar |
[10] Avena, C.V. (2016) Deconstructing the bat skin microbiome: influences of the host and the environment. Front. Microbiol. 7, 1753.
| Deconstructing the bat skin microbiome: influences of the host and the environment.Crossref | GoogleScholarGoogle Scholar |
Biography
Kristin Mühldorfer is a scientist in the Department of Wildlife Diseases at the Leibniz Institute for Zoo and Wildlife Research in Berlin, Germany. She is a certified veterinary microbiologist and head of the bacteriology laboratory. She conducts research and teaches in the field of wildlife microbiology and infectious diseases. Her main interests are in bacteria and fungi of captive and free-living wildlife species from disease ecology, evolutionary adaptation and conservation points of view.


