Cellular signalling by SARS-CoV-2 spike protein
Nicholas P. Gracie A # , Lachlan Y. S. Lai
A # , Lachlan Y. S. Lai  A # and Timothy P. Newsome
A # and Timothy P. Newsome  A B *
A B *
A
B

Nicholas Gracie is a 3rd Year PhD candidate in the Newsome Lab at The University of Sydney. His research involves poxviruses, SARS-CoV-2 and cell signalling. |

Lachlan Lai is a 1st Year PhD candidate in the Newsome Lab at The University of Sydney. His research involves poxviruses with a focus on Monkeypox. |

Timothy Newsome is an Associate Professor of The University of Sydney. His research interests lie at the intersection of virology and cell signalling. |
Abstract
Following the release of the SARS-CoV-2 genome, the spike protein was identified as the key viral protein mediating cell entry. In addition to its critical function in delivering the viral genome to the host cytoplasm, the spike protein is able to activate diverse cell signalling pathways, leading to notable cellular responses, including inflammation, cellular remodelling, and immune evasion. The spike protein is associated with the induction of a ‘cytokine storm’ characterised by elevated levels of proinflammatory cytokines like IL-6 and IL-1β. Moreover, the spike protein deregulates TGF-β and E-selectin, leading to fibrotic injury and tissue scarring in cellular remodelling, notably in pulmonary tissues. Finally, the spike protein plays a role in immune evasion, disrupting Type I interferon responses. Understanding these diverse interactions and effects is crucial for comprehending the pathogenesis of COVID-19 and developing effective therapeutic strategies.
Keywords: ACE2, Angiotensin Converting Enzyme Receptor 2, cellular signalling, COVID-19, cytokine storm, fibrosis, spike protein, TGF-β, transforming growth factor-beta.
SARS-CoV-2, the viral cause of COVID-19, remains a global threat to human health. The spike protein is a glycoprotein encoded by SARS-CoV-2 that mediates membrane fusion and entry into host cells primarily by binding to ACE2. ACE2 expression is highly restricted to specific cell types and largely defines cells that are susceptible to infection although alternative ACE2 pathways have been described.1–4 Current vaccination strategies are also based on delivery of the spike protein through various technologies, which have proven highly effective at reducing transmission, infection, hospitalisation and mortality.5–7 Beyond cell entry and adaptive immune priming, the spike protein also modulates various host cell signalling pathways which ultimately contributes to the pathology and spread of the virus (Fig. 1). These effects can be seen as ACE2-dependent or ACE2-independent, suggesting the spike protein has roles outside of binding. For example, the spike protein is sufficient to induce highly inflammatory cytokines, which may contribute to the multisystem inflammatory syndrome, otherwise known as a ‘cytokine storm’ observed in COVID-19. In addition, the spike protein mediates key changes to the cellular microenvironment that contribute to fibrotic and vascular pathologies. Emerging evidence suggests the spike protein also plays a critical role in subverting host immune signalling to avoid host defences. This review will examine functions of the spike protein independent of ACE2-mediated entry, and how these contribute to COVID-19 pathology.
Different spike protein subunits activate different forms of cell signalling. The spike protein is presented as a trimer on the surface of SARS-CoV-2 virions. The spike protein itself is divided into two subunits, S1 and S2, with S1 encompassing the Receptor Binding Domain (RBD), which initiates ACE2-dependent cell entry, and S2 containing the fusion peptide required for fusion with the host cell membrane. The furin-protease site located between the subunits is unique to SARS-CoV-2, absent in other viruses in the same clade.11 Both full-length spike protein and its component subunits modulate different cell signalling pathways with diverse effects.
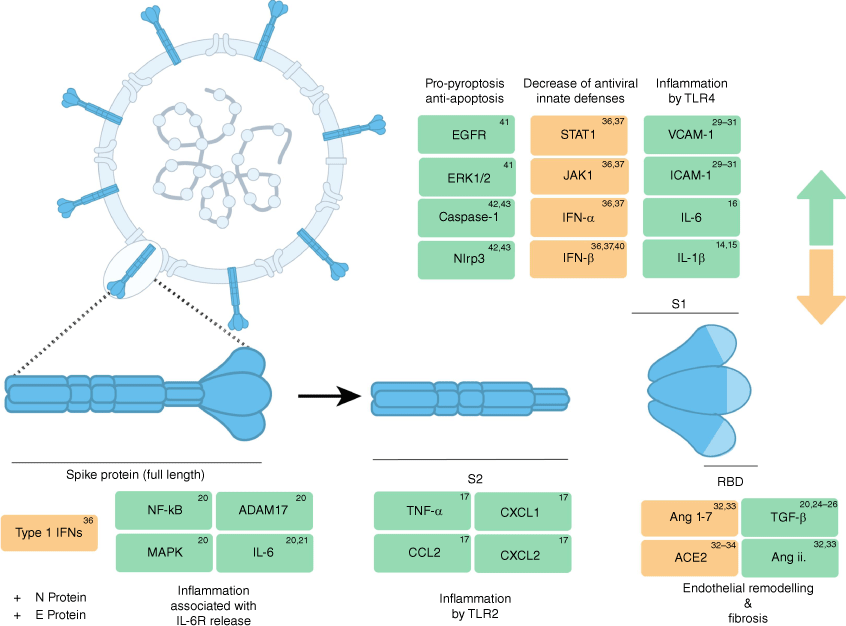
It is important to note that during infection, various subunits and forms of the spike protein can be released and presented to host cells to elicit differing effects (Fig. 2). The spike protein resides on the surface of SARS-CoV-2 virions as trimers. Spike-protein-mediated entry requires two critical cleavage events. Firstly, prior to infection and binding to ACE2, the spike protein is cleaved into subunits S1 and S2 by furin-like proteases. Subsequently, the cleavage of S2′ subunit by the cellular serine protease TMPRSS2, which results in delivery of the viral genome into the cytoplasm.3,8–10 While the spike protein’s role in entry by ACE2 has been extensively described, the modulation of cellular signalling by the spike protein, and its downstream effects, is an expanding area of interest. Here we descriobe three major cellular responses to the spike protein: inflammation, cellular remodelling and immune evasion.
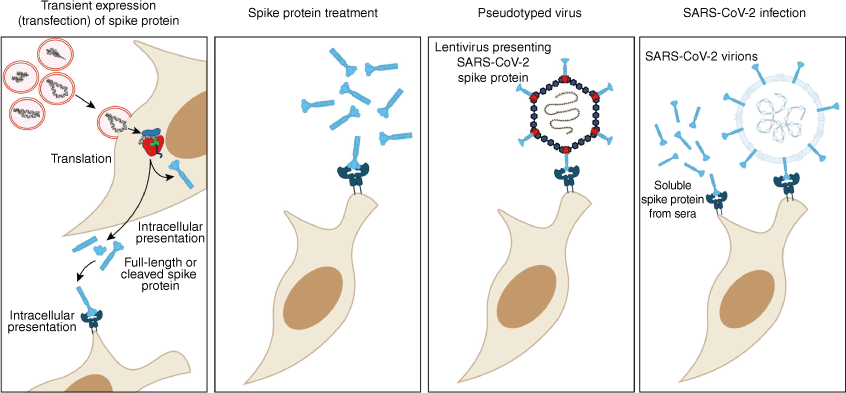
Different modalities for investigating spike-protein activity. Cells are induced to express the spike protein in vitro by transfection or in vivo by mRNA nanoparticle complexes and adenovirus vectors. Post protein synthesis, spike protein forms trimers cleaved at the furin-protease cleavage site between S1 and S2 subunits.23 Synthetic spike protein treatment and pseudotyped virus infection replicate the spike protein binding with ACE2. Live viral infection presents the spike protein to the cell surface and the intracellular environment during virus replication. Additionally, soluble spike protein is present in patients’ sera for up to 12 months post-infection.24
Inflammation
In COVID-19, various proinflammatory cytokines and markers of tissue damage are elevated and associated with severe disease, infection complications and mortality. Such cytokines include, but are not limited to, IL-1β, IL-6, IL-10, TNF, and IFN-ɣ.12 Secretion of proinflammatory IL-1β, IL-12 and TNF is mediated through the Pattern Recognition Receptor (PRR) Toll-like Receptor 4 (TLR4) and NF-κB.12 Recent studies have identified that spike protein in isolation can bind to TLR4 and activate downstream NF-κB signalling, and ultimately drive expression of inflammatory effector genes.13 Purified spike protein trimers directly bind TLR4 and induce IL-1β expression in a dose-dependent manner.14 The spike protein S1 subunit interacts with TLR4, and in conjunction with IFN-ɣ, drives expression of proinflammatory IL-1β to induce the differentiation of proinflammatory M1 macrophages in vitro.15,16 Additionally, spike protein S1 subunit directly interacts with the Leucine-rich repeat domain of TLR4, and in a mouse model of cardiac infection, S1 subunit increased expression of IL-1β and IL-6 to induce inflammation.17 A subsequent study identified a role for the spike protein S2 subunit in the induction of proinflammatory cytokines IL-6, IL-1β, TNF-ɑ and chemokines CXCL1, CXCL2 and CCL2 in human and murine macrophages.18 Here, TLR signalling was activated by the spike protein in TLR4 knockout macrophages, but not TL2 knockouts, suggesting a conflicting dependence on TLR2 rather than TLR4, contrary to previous studies.18
IL-6 has emerged as a key cytokine in driving a hyper-inflammatory state during COVID-19, especially in severe or complicated disease.12 Although clinical trials have failed to show improvement from the use of IL-6 antagonists, it remains evident that the cytokine plays a key role in the development and progression of disease.19 Administration of both polyI:C and the spike protein into ACE2-expressing mice found that IL-6 was induced nearly 100 fold.20 Accordingly, transfecting the spike gene into epithelial cells results in induction of phosphorylated NF-κB, MAPK and secretion of IL-6.21 Importantly, IL-6 trans-signalling may be a key contributor to inflammation-related pathologies.22 During trans-signalling, soluble IL-6 receptors bind IL-6, forming a complex with gp130 that ultimately allows cells not expressing the IL-6 receptor (IL-6R) to respond to IL-6.22 The spike protein has also been shown to activate the ADAM-17 protease, thereby releasing soluble IL-6R, which may increase IL-6 and IL-6R levels in COVID-19.21 Furthermore, an in vitro trans-signalling model demonstrated that exposure to culture fluid from epithelial cells transfected with the spike protein was sufficient to induce IL-6 signalling in endothelial cells, which do not express transmembrane IL-6R.21
Taken together, these studies highlight the role of the spike protein in triggering rapid induction of inflammatory effector genes and signalling molecules that are hallmarks of COVID-19 and correlate with disease severity.
Cellular remodelling
Major complications of COVID-19 include fibrotic injury and tissue scarring. Transforming growth factor-β (TGF-β) is considered the master regulator of fibrosis and tissue remodelling in both homeostasis and disease. In the context of COVID-19, TGF-β, along with multiple other pro-fibrotic cytokines, serves as a biomarker for lung injury.25–27
TGF-β has also been observed to cause endothelial barrier dysfunction following both full-length spike protein and the spike Receptor Binding Domain (RBD) treatment.28 This spike-protein-induced barrier hyperpermeability, an effect observed previously with flavivirus non-structural protein 1 (NS1), has been speculated to promote viral dissemination and pathogenesis.29 Furthermore, it was observed that spike-protein-induced endothelial barrier dysfunction in vitro and vascular leakage in vivo, driven by an upregulation in glycosaminoglycans (GAGs), integrin and TGF-β signalling.28 Cellular remodelling in this study activated by the spike protein was also independent of ACE2. Pulmonary endothelial cell damage in COVID-19 is also tied to paracrine cell signalling. Similar to TGF-β, S1 subunit treatment activates VCAM-1 and ICAM-1 pathways, which contribute to cell vasculopathy. Spike-protein induction of VCAM-1 and ICAM-1 leads to increased expression of E-selectin and risk of blood clotting.30–32
Apart from its role as the key mediator for entry, ACE2 contributes to spike-protein-associated vascular remodelling and fibrosis. As a part of the renin–angiotensin system (RAS), ACE2 controls the hydrolysation of the peptide angiotensin ii to the less inflammatory and pro-fibrotic angiotensin(s) 1–7 during homeostasis. However, this balance is disrupted during vasoconstriction and fibrotic injury.33 In the case of the S1 subunit interaction with ACE2 during viral entry, ACE2 is downregulated, generating an imbalance in the levels of angiotensin ii and leading to an inflammatory response, oxidative stress, vasoconstriction and fibrotic activity.34,35 Similar to live viral infection, multiple methods of presenting the spike protein to cells, including full-length spike protein, ectopic expresison of S1 subunit, and pseudotyped virus infection, are shown to decrease ACE2 expression.34,35 Notably, differences in ACE2 supression were observed between the two studies. Spike-protein treatment reduced ACE2 expression in vitro whereas both treatment and pseudotype virus-expression of the spike protein provided the same effect in vivo.34,35 Additionally, angiotensin ii blockers increase viral replication in SARS-CoV-2 susceptible cell lines, presumably due to an increase in available ACE2.36
Immune evasion
The history of SARS-CoV-2 variants highlight the spike protein as the critical target in immune recognition and driver of enhanced transmissibility and infectivity. In addition, it is becoming increasingly evident that the spike protein itself is able to suppress host-cell Type I Interferon antiviral responses. Specifically, S1 subunit treatment activates the signal transducer and activator of transcription 1 (STAT1) to block association with Janus Kinase 1 (JAK1), leading to reduced IFN-α and IFN-β expression.37,38 A separate study demonstrated that spike and membrane proteins cooperatively decreased IFN-mediated activation of NK cells while enhancing TGF-β activity.39,40
Ectopic expression of the spike protein reduces JAK-STAT activation of IFN activity in cells and spike gene transfection specifically blocks the RIG-1-induced activation of IFN-β with the aid of N protein.41 Associated with this finding, cells transfected with the spike gene led to an increased susceptibility to infection by other RNA viruses.37 IFN-α and IFN-β levels are characteristically low in severe COVID-19 patients compared to mild cases and the activity of spike protein to deregulate IFN production may explain this observation.39
Epidermal Growth Factor Receptor-Mediated (EGFR) signalling, a pathway known to crosstalk with TGF-β, is upregulated following treatment with S1. EGFR signalling also activates ERK1/2 and AKT kinases and enhances the expression of Survivin, a critical inhibitor of apoptosis.42 Further, the spike protein exhibits anti-apoptotic activity in hematopoietic stem cells, triggering Caspase-1 and Nlrp3 and leading to a hyperinflammatory and pyroptotic state.43,44
Discussion
The spike protein has gained much attention for its role in cell entry and as a target of neutralising immunity. Here, we discuss entry-adjacent roles of Spike in initiating an inflammatory ‘cytokine storm’, triggering barrier dysfunction, and silencing host immunity to promote and sustain infection.
Multiple experimental modalities have been used to explore the spike protein as a functional ligand, sometimes with contrasting results. These treatments are often linked to different stages of SARS-CoV-2 infection, where the spike protein can either be presented upon entry into cells, intracellularly during protein synthesis, or in sera from infected patients as its constituent subunits. The spike protein and its subunits can be present extracellularly and intracellularly during and post-infection, which may affect how a cell responds to this ligand. One study found that in patients with post-acute sequelae, spike protein antigen can persist for upwards of 12 months.45 By contrast, the spike protein is not detected in the serum of patients 9 days following primary vaccination, and is undetectable following a secondary vaccination.
Furthermore, different subunits of the spike protein themselves are linked to specific signalling effects, but it remains unknown how each component affects signalling and whether they act independently or cooperatively. Additionally, the cellular machinery required to respond the spike protein or subunits to mediate non-entry function has not been defined. For example, ACE2 is inessential for induction of TGF-β.28 It is therefore highly plausible that different cell types will exhibit a range of responses, and may exceed the cell population susceptible to infection.
We are clearly only beginning to uncover the mechanisms by which the spike protein can deregulate cellular signalling pathways. Here we have reviewed roles for the spike protein in addition to its primary function in mediating cell entry and how these activities may affect COVID-19 pathology. We also highlight that cells can be exposed to the spike protein through various methods and forms, and that these may have distinct disease implications.
References
1 Barthe M et al. (2023) Receptors and cofactors that contribute to SARS-CoV-2 entry: can skin be an alternative route of entry? Int J Mol Sci 24, 6253.
| Crossref | Google Scholar | PubMed |
2 Hikmet F et al. (2020) The protein expression profile of ACE2 in human tissues. Mol Syst Biol 16, e9610.
| Crossref | Google Scholar | PubMed |
3 Jackson CB et al. (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23, 3-20.
| Crossref | Google Scholar | PubMed |
4 Lim S et al. (2022) ACE2-independent alternative receptors for SARS-CoV-2. Viruses 14, 2535.
| Crossref | Google Scholar | PubMed |
5 Martínez-Baz I et al. (2023) Risk reduction of hospitalisation and severe disease in vaccinated COVID-19 cases during the SARS-CoV-2 variant Omicron BA.1-predominant period, Navarre, Spain, January to March 2022. Eurosurveillance 28, pii=2200337.
| Crossref | Google Scholar | PubMed |
6 Andrews N et al. (2022) Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N Engl J Med 386, 1532-1546.
| Crossref | Google Scholar | PubMed |
7 Grgič Vitek M et al. (2022) mRNA vaccine effectiveness against hospitalisation due to severe acute respiratory infection (SARI) COVID-19 during Omicron variant predominance estimated from real-world surveillance data, Slovenia, February to March 2022. Eurosurveillance 27, pii=2200350.
| Crossref | Google Scholar |
8 Cevik M et al. (2020) Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ 371, 3862.
| Crossref | Google Scholar | PubMed |
9 Hoffmann M et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271-280.e8.
| Crossref | Google Scholar | PubMed |
10 Zhang J et al. (2021) Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 372, 525-530.
| Crossref | Google Scholar | PubMed |
11 Coutard B et al. (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176, 104742.
| Crossref | Google Scholar | PubMed |
12 Hsu R-J et al. (2022) The role of cytokines and chemokines in severe acute respiratory syndrome coronavirus 2 infections. Front Immunol 13, 832394.
| Crossref | Google Scholar |
13 Halajian EA et al. (2022) Activation of TLR4 by viral glycoproteins: a double-edged sword? Front Microbiol 13, 1007081.
| Crossref | Google Scholar | PubMed |
14 Zhao Y et al. (2021) SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res 31, 818-820.
| Crossref | Google Scholar | PubMed |
15 Aboudounya MM et al. (2021) SARS-CoV-2 spike S1 glycoprotein is a TLR4 agonist, upregulates ACE2 expression and induces pro-inflammatory M1 macrophage polarisation. bioRxiv 2021.08.11.455921 [Preprint, published 11 August 2021].
| Crossref | Google Scholar |
16 Shirato K, Kizaki T (2021) SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon 7, e06187.
| Crossref | Google Scholar | PubMed |
17 Negron SG et al. (2021) Selectively expressing SARS-CoV-2 spike protein S1 subunit in cardiomyocytes induces cardiac hypertrophy in mice. bioRxiv 2021.06.20.448993 [Preprint, published 20 June 2021].
| Crossref | Google Scholar |
18 Khan S et al. (2021) SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. eLife 10, e68563.
| Crossref | Google Scholar |
19 Jones SA, Hunter CA (2021) Is IL-6 a key cytokine target for therapy in COVID-19? Nat Rev Immunol 21, 337-339.
| Crossref | Google Scholar | PubMed |
20 Gu T et al. (2020) Cytokine signature induced by SARS-CoV-2 spike protein in a mouse model. Front Immunol 11, 621441.
| Crossref | Google Scholar | PubMed |
21 Patra T et al. (2020) SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog 16, e1009128.
| Crossref | Google Scholar | PubMed |
22 Rose-John S et al. (2023) Targeting IL-6 trans-signalling: past, present and future prospects. Nat Rev Immunol 23, 666-681.
| Crossref | Google Scholar | PubMed |
23 Heinz FX, Stiasny K (2021) Distinguishing features of current COVID-19 vaccines: knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines 6, 104.
| Crossref | Google Scholar | PubMed |
24 Swank Z et al. (2023) Persistent circulating severe acute respiratory syndrome coronavirus 2 spike is associated with post-acute coronavirus disease 2019 sequelae. Clin Infect Dis 76, e487-e490.
| Crossref | Google Scholar | PubMed |
25 Karadeniz H et al. (2022) The prognostic value of lung injury and fibrosis markers, KL-6, TGF-β1, FGF-2 in COVID-19 patients. Biomark Insights 17, 117727192211354.
| Crossref | Google Scholar |
26 Willis BC et al. (2005) Induction of epithelial–mesenchymal transition in alveolar epithelial cells by transforming growth factor-β1. Am J Pathol 166, 1321-1332.
| Crossref | Google Scholar | PubMed |
27 Zhang T et al. (2020) Comparison of clinical and pathological features between severe acute respiratory syndrome and coronavirus disease 2019. Zhonghua Jie He He Hu Xi Za Zhi 43, 496-502.
| Crossref | Google Scholar | PubMed |
28 Biering SB et al. (2022) SARS-CoV-2 spike triggers barrier dysfunction and vascular leak via integrins and TGF-β signaling. Nat Commun 13, 7630.
| Crossref | Google Scholar |
29 Biering SB et al. (2021) Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction. Science 371, 194-200.
| Crossref | Google Scholar | PubMed |
30 Kumar N et al. (2021) SARS-CoV-2 spike protein S1-mediated endothelial injury and pro-inflammatory state is amplified by dihydrotestosterone and prevented by mineralocorticoid antagonism. Viruses 13, 2209.
| Crossref | Google Scholar | PubMed |
31 Jana S et al. (2021) Cell-free hemoglobin does not attenuate the effects of SARS-CoV-2 spike protein S1 subunit in pulmonary endothelial cells. Int J Mol Sci 22, 9041.
| Crossref | Google Scholar | PubMed |
32 Meyer K et al. (2021) SARS-CoV-2 spike protein induces paracrine senescence and leukocyte adhesion in endothelial cells. J Virol 95, e00794-21.
| Crossref | Google Scholar | PubMed |
33 Fountain JH et al. (2023) Physiology, Renin Angiotensin System. StatPearls Publishing, Treasure Island, FL, USA. https://www.ncbi.nlm.nih.gov/books/NBK470410/
34 Gao X et al. (2022) Spike-mediated ACE2 down-regulation was involved in the pathogenesis of SARS-CoV-2 infection. J Infect 85, 418-427.
| Crossref | Google Scholar | PubMed |
35 Lei Y et al. (2021) SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE 2. Circ Res 128, 1323-1326.
| Crossref | Google Scholar | PubMed |
36 Pires De Souza GA et al. (2021) Angiotensin II receptor blockers (ARBs antihypertensive agents) increase replication of SARS-CoV-2 in Vero E6 cells. Front Cell Infect Microbiol 11, 639177.
| Crossref | Google Scholar | PubMed |
37 Sui Y et al. (2021) SARS-CoV-2 spike protein suppresses ACE2 and type I interferon expression in primary cells from macaque lung bronchoalveolar lavage. Front Immunol 12, 658428.
| Crossref | Google Scholar |
38 Zhang Q et al. (2021) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) and spike (S) proteins antagonize host type i interferon response. Front Cell Infect Microbiol 11, 766922.
| Crossref | Google Scholar |
39 Csordas BG et al. (2022) Is IFN expression by NK cells a hallmark of severe COVID-19? Cytokine 157, 155971.
| Crossref | Google Scholar | PubMed |
40 Ferreira-Gomes M et al. (2021) SARS‐CoV‐2 in severe COVID‐19 induces a TGF‐β‐dominated chronic immune response that does not target itself. Nat Commun 12, 1961.
| Crossref | Google Scholar |
41 Freitas RS et al. (2022) SARS-CoV-2 spike antagonizes innate antiviral immunity by targeting interferon regulatory factor 3. Front Cell Infect Microbiol 11, 789462.
| Crossref | Google Scholar | PubMed |
42 Palakkott AR et al. (2023) The SARS-CoV-2 spike protein activates the epidermal growth factor receptor-mediated signaling. Vaccines 11, 768.
| Crossref | Google Scholar | PubMed |
43 Kucia M et al. (2021) An evidence that SARS-Cov-2/COVID-19 spike protein (SP) damages hematopoietic stem/progenitor cells in the mechanism of pyroptosis in Nlrp3 inflammasome-dependent manner. Leukemia 35, 3026-3029.
| Crossref | Google Scholar | PubMed |
44 Ratajczak MZ et al. (2021) SARS-CoV-2 entry receptor ACE2 is expressed on very small CD45− precursors of hematopoietic and endothelial cells and in response to virus spike protein activates the Nlrp3 inflammasome. Stem Cell Rev Rep 17, 266-277.
| Crossref | Google Scholar | PubMed |
45 Ogata AF et al. (2022) Circulating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccine antigen detected in the plasma of mRNA-1273 vaccine recipients. Clin Infect Dis 74, 715-718.
| Crossref | Google Scholar | PubMed |
 Nicholas Gracie is a 3rd Year PhD candidate in the Newsome Lab at The University of Sydney. His research involves poxviruses, SARS-CoV-2 and cell signalling. |
 Lachlan Lai is a 1st Year PhD candidate in the Newsome Lab at The University of Sydney. His research involves poxviruses with a focus on Monkeypox. |
 Timothy Newsome is an Associate Professor of The University of Sydney. His research interests lie at the intersection of virology and cell signalling. |


