The evaluation of web-based data collection for enhanced surveillance of cryptosporidiosis
Kerri A. Viney A C and Jeremy M. McAnulty BA NSW Public Health Officer Training Program, NSW Department of Health
B Communicable Diseases Branch, NSW Department of Health
C Corresponding author. Email: kvine@doh.health.nsw.gov.au
NSW Public Health Bulletin 19(2) 15-19 https://doi.org/10.1071/NB07103
Published: 28 February 2008
Abstract
Following an increase in the number of people diagnosed with cryptosporidiosis in November 2005, the Communicable Diseases Branch initiated enhanced surveillance using a developmental version of NetEpi Collection, an open-source, web-based data collection tool. We evaluated the usefulness of NetEpi Collection for enhanced surveillance, using the Centers for Disease Control and Prevention’s Updated Guidelines for Evaluating Public Health Surveillance Systems as a guide. Most staff (73 per cent) who used NetEpi Collection found it easy to use. Although ongoing support was thought to be adequate by 82 per cent of respondents who used NetEpi Collection, 36 per cent reported that training was limited and 27 per cent reported technical problems such as internet, server and password problems. In order to improve its usefulness in enhanced surveillance, training in NetEpi Collection should be enhanced and the stability of the system improved.
Cryptosporidiosis is a diarrhoeal disease caused by the intestinal parasite, Cryptosporidium.1 Outbreaks are common and have been associated with swimming pools, drinking water supplies and, rarely, consumption of contaminated beverages.1 Under the NSW Public Health Act 1991, cryptosporidiosis is a notifiable disease and must be reported to NSW Health by laboratories. In NSW, eight public health units (PHUs) located across 16 sites are responsible for following up notifications of communicable diseases including cryptosporidiosis. PHU staff enter cases of cryptosporidiosis onto the state-wide surveillance system, the Notifiable Diseases Database (NDD), and investigate these using a standardised cryptosporidiosis questionnaire.
Establishing enhanced surveillance
In November 2005, analysis of routinely collected surveillance data by the NSW Department of Health identified an increase in the number of people notified with cryptosporidiosis. Following initial anecdotal reports that many of these cases reported contact with farms or cattle, the NSW Department of Health initiated enhanced surveillance to explore and quantify risk factors among the cases.
From 1 November 2005 to 29 May 2006 (the outbreak period), PHU staff were asked to interview all cases of cryptosporidiosis using the standard NSW Health questionnaire. The questionnaire included potential risk factors associated with cryptosporidiosis, including farm visits, preschool attendance, swimming and contact with another case. Cryptosporidiosis cases were entered into NDD in the usual way. However, as NDD does not have the capacity to capture enhanced surveillance data, staff from the Communicable Diseases Branch at the NSW Department of Health designed a database using NetEpi Collection software for this purpose. NetEpi Collection is an open-source, web-based software that has been developed by the Centre for Epidemiology and Research, at the NSW Department of Health, to collect structured information about cases and contacts through web browsers on the internet.2
One PHU staff member from each PHU site was asked to enter additional risk factor data and the NDD case number into the developmental version (version 0.95) of NetEpi Collection. Training was provided to PHU staff via a teleconference, and ongoing individual support was provided using email or telephone. Data from NDD and NetEpi Collection were then merged and analyses performed using SAS (version 8.2) to determine what risk factors were important in ongoing transmission during the outbreak.3
The aim of the present study was to assess the usefulness of a web-based data collection system (such as NetEpi Collection) for enhanced surveillance during a communicable disease outbreak.
Methods
We used the Centers for Disease Control and Prevention’s Updated Guidelines for Evaluating Public Health Surveillance Systems as a framework to:
-
assess the public health importance of the surveillance system
-
assess the usefulness of the surveillance system
-
describe attributes of the surveillance system (i.e. simplicity, flexibility, data quality, acceptability, predictive value positive, representativeness, timeliness and stability).4
Public health importance of cryptosporidiosis
A review of the public health literature was undertaken to assess the outbreak potential and preventability of cryptosporidiosis. NSW cryptosporidiosis data were analysed to compare notifications for the outbreak period with the previous five years (2000–2005).
Description of surveillance system components
Managers of NDD and NetEpi Collection were asked to provide a description of each system component.
Usefulness of the surveillance system
At the end of the outbreak period, one person from each of the 16 PHU sites was asked to complete a questionnaire (Box 1). The questionnaire outlined their experience of using NetEpi Collection, and responses were collated in Excel (version 5.0.2). Risk factor data was analysed at a state level to determine if NetEpi Collection was useful in assessing statewide risk factors associated with cryptosporidiosis.

|
Attributes of the surveillance system
Acceptability
We asked PHU staff if they used NetEpi Collection, whether it was useful or burdensome, and whether there were barriers to using NetEpi Collection in this outbreak. We also asked about the adequacy of training and ongoing support provided by the Communicable Diseases Branch.
Stability
To determine the system’s ability to manage data without failure, PHU staff were asked about any technical difficulties experienced while using NetEpi Collection.
Quality and representativeness of data
A comparison of the cryptosporidiosis data in NetEpi Collection and NDD was used to determine the completeness of Net Epi data and whether it was representative of all cases. This allowed us to determine which PHUs entered data into NetEpi Collection and what was different about the cases that were not entered. We also examined the completeness of data in the risk factor fields in NetEpi Collection.
Results
Public health importance of cryptosporidiosis
In NSW and elsewhere, cryptosporidiosis has been linked to swimming in contaminated swimming pools.5–7 Although outbreaks associated with drinking water supplies are rare in Australia, there is the potential for large outbreaks due to contaminated water sources, such as the Milwaukee outbreak of 1993.8,9 Early detection of cryptosporidiosis outbreaks may help public health attempts to develop interventions aimed at preventing further disease.
Between 1997–2006, the number of cryptosporidiosis notifications in NSW has ranged from 1130 in 1998 to 121 in 1999, with an average of 423 cases per year. In the outbreak period, a total of 871 cases of cryptosporidiosis were reported to NSW Health. During the same time period in the previous five years, there was average of 235 cases of cryptosporidiosis reported each year.
Components of the surveillance system
Surveillance of cryptosporidiosis begins when a person with symptoms visits a doctor, and a faecal specimen is ordered and sent to a laboratory for testing. If the result is positive, the laboratory notifies the local PHU where the case is entered into the NDD and the person interviewed for risk factors. PHU staff review these data to identify clustering of cases (time and location) and, where necessary, initiate control measures following the NSW Health Cryptosporidiosis Response Protocol for NSW Public Health Units.10 Data are then available for analysis by the Communicable Diseases Branch, and reports are compiled for the NSW Health Public Health Bulletin and NSW Health website.11,12
The NDD is a secure, decentralised database used for storing information on cases of notifiable diseases in NSW.13 Cryptosporidiosis data entered into NDD includes demographic variables, disease characteristics, disease outcome, laboratory information, organism, specimen type, identification method and other variables. Risk factors are not entered into NDD, but NDD does contain a ‘clinical notes’ text field, where risk factors for disease may be entered.
During the outbreak period, specific risk factor information was entered into the NetEpi Collection database. This database contained a unique identifier common to NDD and NetEpi Collection, as well as fields on risk factors for cryptosporidiosis such as: swimming; contact with another case; preschool attendance; and farm visit. PHU staff entered the common identifier and risk factor data into the NetEpi Collection database after entering the case into NDD in the usual manner.
Usefulness of the surveillance system
Just over two-thirds of PHU sites reported using NetEpi Collection during the outbreak. Of those who used NetEpi Collection, most found it easy to use (Table 1). Just under one-third of those who used NetEpi Collection exported data for analysis and the data were infrequently used to assess local risk factors associated with cryptosporidiosis.

|
At the statewide level, Communicable Diseases Branch staff thought that NetEpi Collection provided useful information on risk factors, in particular the geographic distribution and timing of exposures.14 Analysis of statewide data entered into NetEpi Collection revealed that the outbreak may have started in rural areas of NSW with farm animal contact as a risk factor. The outbreak then appeared to involve spread by contact to other parts of NSW, with person-to-person contact and swimming pool contamination becoming more important as risk factors.14
Attributes of the surveillance system
Acceptability
Of the 11 PHU staff who used NetEpi Collection, 9 (82 per cent) found it useful because it provided: an improvement of surveillance; an analysis of disease trends; and assistance in determining clusters and risk factors. Time was a factor for those who found the system burdensome, and barriers to using NetEpi Collection for analysis ranged from technical difficulties to lack of training (Table 1).
While training was perceived to be adequate by more than half of the users, most reported that they did not analyse the data collected through NetEpi Collection (Table 1). Two people who thought that training was not adequate were unable to attend the initial training session.
Stability
PHU staff experienced technical difficulties using NetEpi Collection (Table 1). These included: difficulty with accessing the internet; occasional server/program crashes; slow internet connection; and an inability to use the return key to move between data entry fields (the tab key needs to be used instead). Some respondents mentioned that problems had been fixed after liaison with staff from the Communicable Diseases Branch.
Quality and representativeness of data
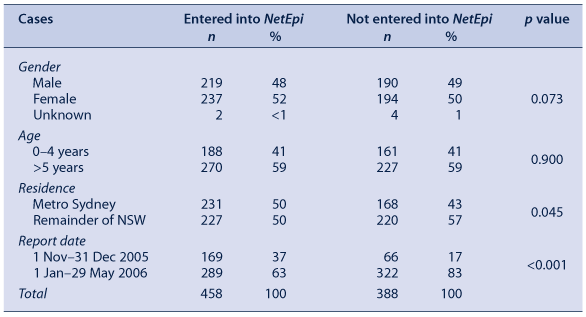
At June 2007, a total of 846 cases of cryptosporidiosis were reported for the outbreak period (Table 2). Of these, 458 (54 per cent) were entered into NetEpi Collection (Table 2). In the risk factor fields in NetEpi Collection, the data fields were, on average, 86 per cent complete.
|
Lack of time meant that it was not possible to enter data into NetEpi Collection at all PHU sites (5/16 or 31 per cent). Cases entered into NetEpi Collection were similar to the NDD cases with respect to gender and age (p = 0.073 and 0.900 respectively) (Table 2). A larger proportion of cases from metropolitan Sydney were entered into NetEpi Collection than from other parts of NSW (p = 0.045) and a higher proportion were entered at the beginning of the outbreak than later in the outbreak (p = 0.000).
Discussion
In an outbreak setting, enhanced surveillance of cryptosporidiosis using a web-based data collection system allowed identification of risk factors for disease that could not have been achieved using NDD alone. Information describing risk factors also allowed identification of area of residence of those potential exposures over time.
PHU staff infrequently exported and used NetEpi Collection data to assess local risk factors for transmission. No comparison can be made with using the NDD system in this way. Nevertheless, despite time and technological limitations, most PHU staff thought that using enhanced surveillance would help in determining clusters and risk factors.
The information entered into NetEpi Collection was useful for analysing risk factors at a statewide level. Such web-based data collection systems may offer greater advantages for the co-ordinating site, as they enable analysis of risk factor information for the whole dataset. The use of web-based data collection tools may also be a more streamlined method of data collection than other methods (such as faxing or emailing single databases to a central point) and so could prevent time delays and errors in data transcription. However, where broadband access is unreliable or slow, the use of web-based data collection tools may be limited.
The observed decreasing compliance with entering data into NetEpi Collection suggests that enhanced surveillance may be more sustainable over short periods.
To improve the use of web-based data collection tools, both training and support with technical problems should be improved. This could include repeating introductory training sessions and providing training in data analysis. In addition, PHU sites require adequate and reliable access to the internet.
With regards to the system itself, the stability needs to be improved and the data entry interface should allow staff to use the return key to move between fields when entering data. The authors of NetEpi Collection have advised that version 1.0 of the software, which includes numerous enhancements, will be publicly available at http://www.netepi.org when this paper is published.
In previous outbreak investigations carried out by NSW Health, risk factor information has been collected by faxing forms to a central point, then entering into a database. Elsewhere in Australia, Canada and the United Kingdom, enhanced surveillance has been used to collect risk factor data for a range of diseases in outbreak situations. This approach has been recently applied to the analysis of risk factors for hepatitis B,15,16 meningococcal disease,17 haemolytic uraemic syndrome,18 hepatitis C19 and campylobacter,20 among other diseases. OzFoodNet have used an even earlier developmental version of NetEpi Collection during an outbreak of Salmonella Hvittingfoss in order to identify potentially implicated foods and the investigators concluded that using a web-based data collection system such as NetEpi Collection was a dramatic improvement in the collection of data in a geographically dispersed outbreak.21
Conclusion
Enhanced surveillance of cryptosporidiosis provided useful information on potential exposures during an outbreak with widespread geographic distribution. Using the web-based data collection tool, NetEpi Collection, made analysis of data easier for the co-ordinating site, but was difficult and time-consuming for some PHU staff.
Where systems data are complete and representative, enhanced surveillance during outbreaks using web-based data collection tools can provide useful information about exposures for disease.
Acknowledgements
Many thanks to Tim Churches (Population Health Information Branch, NSW Health) for advice on the manuscript and to the Public Health Unit network for their co-operation during the outbreak and in returning questionnaires.
[1]
[2]
[3]
[4] Centers for Disease Control and Prevention Updated guidelines for evaluating public health surveillance systems: Recommendations from the Guidelines Working Group. MMWR 2001; 50 1–36.
| PubMed |

[5] Puech MC, McAnulty JM, Lesjak M, Shaw N, Heron L, Watson JM. A statewide outbreak of cryptosporidiosis in New South Wales associated with swimming at public pools. Epidemiol Infect 2001; 126(3): 389–96.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[6] Stafford R, Neville G, Towner C. McCall B. A community outbreak of Cryptosporidium infection associated with a swimming pool complex. Commun Dis Intell 2000; 24(8): 236–9.
| PubMed |

[7] Veverka F, Shapiro N, Parish MK, York S, Becker W, Smith F, et al. Protracted outbreaks of cryptosporidiosis associated with swimming pool use-Ohio and Nebraska, 2000. MMWR 2001; 50(20): 406–10.
| PubMed |

[8] Lester R. A mixed outbreak of cryptosporidiosis and giardiasis. Quarterly Bull Infect Dis 1992; 1(1): 14–15.

[9] Mac Kenzie WR, Hoxie NJ, Proctor ME, Gradus MS, Blair KA, Peterson DE, et al. A massive outbreak in Milwaukee of cryptosporidium infection transmitted through the public water supply. N Engl J Med 1994; 331(3): 161–7.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[10]
[11]
[12]
[13] Persson L, Bartlett M. Notifiable Diseases Database System: review and development strategy. N S W Public Health Bull 2004; 15(1–2): 10–12.
| PubMed |

[14] Viney K, McAnulty J. Statewide outbreak of cryptosporidiosis in NSW, November 2005–May 2006. N S W Public Health Bull 2007; 18(7–8): 141–4.

[15] Poulos RG, Ferson M. Enhanced surveillance of acute hepatitis B in south-eastern Sydney. Commun Dis Intell 2004; 28(3): 392–5.
| PubMed |

[16] Forssman BL, Gupta L. Enhanced surveillance of Hepatitis B infection in inner Western Sydney. N S W Public Health Bull 2007; 18(7–8): 115–8.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[17] Hahne SJM, Gray SJ, Aguilera J-F, Crowcrift NS, Nichols T, Kaczmarski EB, et al. W135 meningococcal diseae in England and Wales associated with the Hajj 2000 and 2001. Lancet 2002; 359 582–3.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[18]
[19] Wu H-X, Wu J, Wong T, Donaldson T, Dinner K, Andonov A, et al. Enhanced surveillance of newly acquired hepatitis C infection in Canada, 1998 to 2004. Scand J Infect Dis 2006; 38 482–9.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[20] Sopwith W, Ashton M, Frost JA, Tocque K, O’Brien S, Regan M, et al. Enhanced surveillance of Campylobacter infection in the North West of England 1997–1999. J Infect 2003; 46 35–45.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

[21] Oxenford CJ, Black AP, Bell RJ, Munnoch SA, Irwin MJ, Hanson RN. Owen RL and the Outbreak Investigation Team. Investigation of a multi-state outbreak of Salmonella Hvittingfoss using a web-based case reporting form. Commun Dis Intell 2005; 29(4): 379–81.
| PubMed |


