Wastewater, wheat and table wipes: adventures in culture-independent microbiology
Jacob E Munro A , Deborah J Rich A , Simon Dingsdag A and Nicholas V Coleman A BA School of Molecular Bioscience
Building G08, University of Sydney
Darlington, NSW 2006, Australia.
B Corresponding author. Tel: +61 2 9351 6047
Fax: +61 2 9351 5858
Email: nicholas.coleman@sydney.edu.au
Microbiology Australia 35(4) 188-191 https://doi.org/10.1071/MA14061
Published: 30 October 2014
The sequencing of ribosomal RNA and DNA (rRNA/rDNA) from environmental samples heralded a new age in microbiology1–3. The advent of next-generation sequencing supercharged these methods, which now give high-resolution data sets, enabling real insights into microbial diversity and function in complex systems4–7. Here, three local applications of 16S rDNA pyrosequencing are described, which highlight the usefulness of this approach for addressing practical questions in diverse areas of microbiology. Limitations of the sequence-based approach will also be discussed.
Wastewater: understanding the shutdown response in biological aerated filters
An industry partner approached our lab for assistance with managing microbes in the biological aerated filter units (BAFs) in their wastewater treatment plant. The BAFs were designed to degrade volatile fatty acids (VFAs) in the wastewater stream, but were problematic after ‘shutdown’ events. Such events involve stopping the water flow for many days, and in some cases cleaning the BAFs; these actions change the physiology and/or community in the BAFs such that they do not readily re-start VFA oxidation.
We sampled the BAF material (zeolite+biofilm) at intervals over a time course spanning both shutdown and restart events. RNA was extracted, and reverse-transcribed to cDNA, then used for 16S and 18S rRNA gene PCRs, and tag-pyrosequencing. We used bead-beating combined with a commercial RNA extraction kit to good effect; this was an ‘easy’ template due to the abundance of biomass in the BAF material (Figure 1). Our rationale for using ribosomal RNA as the template was that this is a better marker for the activity of microbes compared to ribosomal DNA, which is better correlated to cell abundance (e.g. see Hunt et al.8).
|
Distinctive changes in the BAF community occurred during shutdown (representative data from one of three replicate BAFs are shown in Figure 2); Arcobacter, Zoogloea, and Bdellovibrio declined, while Rubrivivax and Pedomicrobium increased. After restart of flow, the community seemed to return to the initial structure, but this robust response to perturbation at the genus level did not correspond to success in restarting VFA oxidation (data not shown). The changes involved in the shutdown response may be occurring at finer-scale taxonomic resolution, or might not involve ribosome abundance (e.g. they may involve enzyme induction).

|
Bdellovibrio is a predator on other bacteria. This genus suffered dramatic declines in rRNA abundance after shutdown (50- to 150-fold). It is interesting that this ‘top predator’ was the most sensitive to ecosystem disturbance – this mirrors patterns seen in macro-organisms9. Bdellovibrio may be a predator of a bacterium that is inhibitory to VFA oxidation, or it may be acting here as an indicator organism of the chemical changes in the system. Further work is needed to elucidate the microbial basis of the BAF shutdown response.
Wheat: tracking inoculant strains and discovering indigenous microbiota
Ethylene (C2H4) is a gaseous plant hormone10. Ethylene-oxidising bacteria can be readily isolated from soil11 – do these bacteria interact with plants based on the ethylene system? Alternatively, these bacteria could be consuming ethylene made by microbial fermentation12…the jury is out. Ethylene-oxidising isolates are nearly always fast-growing Mycobacterium spp.13,14 – these are a fascinating group of microbes, which are mostly non-pathogenic, but highly immunogenic15. They may even play a role in influencing our moods16,17.
We have begun a study to investigate the interactions of ethylene-oxidising bacteria and plants. The 16S pyrosequencing approach was used to provide information on the persistence of the inoculant strains and to reveal which types of indigenous bacteria were present. Note that although the tag-pyrosequencing data are not quantitative in the sense that sequences do not correlate 1 : 1 with cells, the data can be used to discern trends in relative abundances, and provide a reference point for viable counts in the case of the inoculant strain.
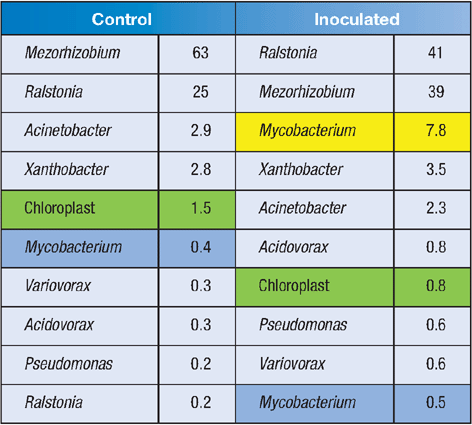
Preliminary data from two individual wheat plants (control/inoculated) are shown in Table 1. Note that the DNA extraction method used here has captured both the surface microbiota and the endophytes in the wheat plants, as evidenced by the abundant chloroplast sequences that are recovered (green highlight). These organelles contain their own 16S rDNA, which bears testament to their cyanobacterial ancestry18. At this stage it is not clear which of these taxa are surface microbiota, and which are endophytes.
|
The tag-pyrosequencing approach easily detected our inoculated ethylene-oxidising bacterium (yellow highlight), but intriguingly, the sequence data also revealed an indigenous Mycobacterium species (blue highlight) – this was the 6th-most abundant bacterial sequence detected in the uninoculated plants. Closer inspection of this sequence reveals that the indigenous Mycobacterium was closely related to species known as ethylene-oxidisers. Is this organism involved in ethylene oxidation in vivo in the wheat plant?
Wipes: assessing risks from bacteria in a shopping centre food court
Our lab was contacted by a union representing cleaners to undertake an investigation into the microbiology of the shopping centre food court. There was concern from the union that the cleaners were under-resourced, based on rumours of poor practices such as re-using cleaning cloths for excessive lengths of time, or using the same cloths in multiple places (e.g. bathrooms and food court).
We obtained samples of a cleaning cloth used in the food court, and also table-wipes from many individual table surfaces. Our aims were to determine the total bacterial numbers (plate count), to determine if pathogenic bacterial types were present, and to determine if faecal indicator organisms (E. coli) were present. These data would allow us to estimate firstly the level of public health risk, and secondly, to see if there was evidence for cross-area usage of cloths between food court and bathroom (for full study details, see Dingsdag19).
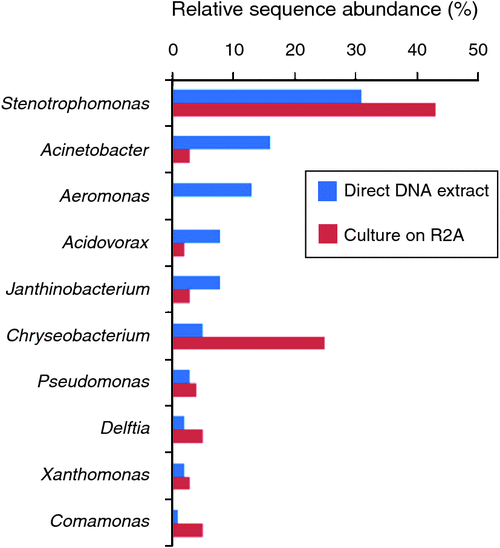
As part of this study, we wanted to get a sense of the relationship between the bacterial types growing on the agar used (R2A), and the total community in the cleaning cloth – are the isolates grown on agar really representative of the major types present in this environment? This once intractable-question is now easy to answer by doing a 16S PCR on DNA extracted direct from the cloth, and another PCR on DNA extracted from the pooled colonies grown on agar, and pyrosequencing both PCR products (Figure 3).
|
The majority of bacterial types detected on agar plates were consistent with those detected by direct DNA extraction from the cleaning cloth, although their relative order changed. Aeromonas was the exception; this was third-most abundant genus in the cloth, but did not grow at all on R2A. This could be due to inhibition by other faster-growing bacteria, since aeromonads can certainly be cultivated on R2A20.
The ease of culturability of most bacteria in the cloth could be because this is a eutrophic environment (like culture media), or perhaps this habitat selects stress-resistant types (detergents, heat); these may also resist the stress of isolation on agar.
The most abundant sequences detected in the cleaning cloth were from genera that include human pathogens, such as Stenotrophomonas, Acinetobacter, and Aeromonas. While tag-pyrosequencing (~400 bp sequence) cannot reliably identify bacteria to the species level, the closest sequence matches in many cases (>99%) were to pathogens such as S. maltophilia, Ac. baumannii and Ae. hydrophila. Both coliforms (Enterobacter, Citrobacter) and faecal coliforms (E.coli) were detected, with E.coli at 0.3–0.5% of sequences – this may indicate the use of this cloth in the bathroom, but could also be due to poor hygiene of the general public, who contribute to the bacterial load here.
Limitations of the tag-pyrosequencing approach
Any type of PCR will be limited by the primers used. The ‘universal’ primers for targeting 16S and 18S rDNA are not identical to all-prokaryote or all-eukaryote ribosomal sequences, respectively, and they cannot amplify all of the sequence types in a complex habitat21. Further, different sequence types will be amplified with differing efficacy, if present in a mixed DNA template. The latter effect is marked, and can be demonstrated by PCR and sequencing of defined mixtures of a few dozen ribosomal sequences22. Another serious problem with PCRs from mixed templates is the generation of chimeric sequences, which need to be specifically detected and removed23.
Biases also arise from nucleic acid extraction, since different microbes are lysed with different efficacy. Physical disruption (e.g. bead-beating) is often the method of choice, since it is rapid, and it can lyse both bacteria and eukaryotes, but neither this method or the alternatives are guaranteed to lyse all microbial types, which leads to a bias towards the more easily lysed types in sequence libraries24,25. The peculiarities of different environmental matrices (e.g. soils vs. clinical samples) impact strongly on the yield and purity of extracted DNA, and its usefulness for downstream amplification steps26. This is a particular challenge for forensic use of tag-pyrosequencing, where legal decisions are made based on sequence data27.
Pyrosequencing is more error-prone than Sanger sequencing, and the level of errors generated can be sufficient to yield false operational taxonomic units (OTUs) if rigorous sequence quality control is not employed. This means that unique clones in sequence data may be genuine members of the rare biosphere, or they may be sequencing errors28,29. Another pyrosequencing artefact is the generation of false clusters of identical or closely related sequences – these are present at up to 35% in some metagenomic datasets30.
Many traps in tag-pyrosequencing analysis relate to over-interpretation of the data31 – this could include extrapolating cell numbers from numbers of rDNA sequences (rRNA gene copy number varies in different phyla), postulating physiological functions based on ribosome sequences (most bacteria have highly variable metabolism and physiology), or mistaking statistical correlation s between sequences as causal linkages (an error in logic).
Concluding remarks
Environmental microbiologists have an important role to play in addressing humanity’s major challenges in the 21st Century. Our technical ability to attack these problems is more powerful than ever, but our efforts locally are constrained by a lack of funding and a lack of vision from our large institutions and governments. As a Society, and as individual microbiologists, we need to push harder for recognition of the reality and seriousness of environmental problems, and the importance of microbiologists in solving these problems.
Acknowledgements
Funding for the Coleman lab has been provided by the Australian Research Council, The NSW Environmental Trust, The University of Sydney, Orica Ltd, Environmental Earth Sciences Inc., Austar Coal, and United Voice.
References
[1] Pace, N.R. (1997) A molecular view of microbial diversity and the biosphere. Science 276, 734–740.| A molecular view of microbial diversity and the biosphere.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXivFOltLc%3D&md5=eb1cf8c1be37b8a8462756d70f82cbadCAS | 9115194PubMed |
[2] Ward, D.M. et al. (1992) Ribosomal RNA analysis of microorganisms as they occur in nature. In Advances in Microbial Ecology (Marshall, K.C., ed), pp. 219–286, Plenum Press, New York, NY, USA; London, England, UK.
[3] Barns, S.M. et al. (1994) Remarkable archaeal diversity detected in a Yellowstone National Park hot spring. Proc. Natl. Acad. Sci. USA 91, 1609–1613.
| Remarkable archaeal diversity detected in a Yellowstone National Park hot spring.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2cXitFanurw%3D&md5=da877e49faf7c35d9a415d3933d8dd68CAS | 7510403PubMed |
[4] Lauber, C.L. et al. (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120.
| Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVSgtrjJ&md5=67ea04b9c9a01e748192981392af7cf2CAS | 19502440PubMed |
[5] Acosta-Martínez, V. et al. (2008) Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 40, 2762–2770.
| Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use.Crossref | GoogleScholarGoogle Scholar |
[6] Koenig, J.E. et al. (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 108, 4578–4585.
| Succession of microbial consortia in the developing infant gut microbiome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjvVCkurs%3D&md5=c35f4c18218146dd71f657ab1bb30c7cCAS | 20668239PubMed |
[7] Tas, N. et al. (2014) Impact of fire on active layer and permafrost microbial communities and metagenomes in an upland Alaskan boreal forest. ISME J. 8, 1904–1919.
| Impact of fire on active layer and permafrost microbial communities and metagenomes in an upland Alaskan boreal forest.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsVWksr7P&md5=3a6b0bc8774cd3d9dcb965670e52b368CAS | 24722629PubMed |
[8] Hunt, D.E. et al. (2013) Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl. Environ. Microbiol. 79, 177–184.
| Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXkvVKjsLs%3D&md5=9b3781817a95f8b966791110864e20adCAS | 23087033PubMed |
[9] Rattner, B.A. (2009) History of wildlife toxicology. Ecotoxicology 18, 773–783.
| History of wildlife toxicology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVeksrnJ&md5=dc0f9cb13bbaf572f76de82f33022065CAS | 19533341PubMed |
[10] Bleecker, A.B. and Kende, H. (2000) Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18.
| Ethylene: a gaseous signal molecule in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXpvFOj&md5=cedcae7da60de455d4583e3f3d43dcf0CAS | 11031228PubMed |
[11] de Bont, J.A. (1976) Oxidation of ethylene by soil bacteria. Antonie van Leeuwenhoek 42, 59–71.
| Oxidation of ethylene by soil bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE28XktlSgsbs%3D&md5=527d6c8e6d52588e289e946d26053f5aCAS | 1085129PubMed |
[12] Fukuda, H. et al. (1993) Ethylene production by micro-organisms. Adv. Microb. Physiol. 35, 275–306.
| Ethylene production by micro-organisms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2cXkslOrsbw%3D&md5=438bcb8cb9ab67446be4259e02bc0011CAS | 8310882PubMed |
[13] Mattes, T.E. et al. (2010) Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol. Rev. 34, 445–475.
| 1:CAS:528:DC%2BC3cXosFChsbw%3D&md5=ee6f00da34f8d48aa2b4efb96cfbc8beCAS | 20146755PubMed |
[14] Coleman, N.V. and Spain, J.C. (2003) Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl. Environ. Microbiol. 69, 6041–6046.
| Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXotlKjsrg%3D&md5=432b918ef79065b8b722fa7c522b1df8CAS | 14532060PubMed |
[15] Fine, P.E.M. (1995) Variation in protection by BCG – implications of and for hetrologous immunity. Lancet 346, 1339–1345.
| Variation in protection by BCG – implications of and for hetrologous immunity.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK28%2FmsV2nsg%3D%3D&md5=00d90182dc6f57d0d1ae9c5aabae7fb1CAS |
[16] Lowry, C.A. et al. (2007) Identification of an immune-responsive mesolimbocortical serotonergic system: Potential role in regulation of emotional behavior. Neuroscience 146, 756–772.
| Identification of an immune-responsive mesolimbocortical serotonergic system: Potential role in regulation of emotional behavior.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXkslajtbg%3D&md5=c70f4a00c6ca8382898849d1d3001194CAS | 17367941PubMed |
[17] Raison, C.L. et al. (2010) Inflammation, sanitation, and consternation loss of contact with coevolved, tolerogenic microorganisms and the pathophysiology and treatment of major depression. Arch. Gen. Psychiatry 67, 1211–1224.
| Inflammation, sanitation, and consternation loss of contact with coevolved, tolerogenic microorganisms and the pathophysiology and treatment of major depression.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjtFyquw%3D%3D&md5=1ac603380fa7e748c7a7f5890f30f403CAS | 21135322PubMed |
[18] Giovannoni, S.J. et al. (1988) Evolutionary relationships among cyanobacteria and green chloroplasts. J. Bacteriol. 170, 3584–3592.
| 1:STN:280:DyaL1c3pvVKlug%3D%3D&md5=df7c6879701407778acab1483d061abfCAS | 3136142PubMed |
[19] Dingsdag, S. and Coleman, N.V. (2013) Bacterial communities on food court tables and cleaning equipment in a shopping mall. Epidemiol. Infect. 141, 1647–1651.
| Bacterial communities on food court tables and cleaning equipment in a shopping mall.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38bos1GktA%3D%3D&md5=0a545b8d8b6a24068ec70fc8d2f88a0eCAS | 22995219PubMed |
[20] Kampfer, P. et al. (1996) Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb. Ecol. 32, 101–121.
| Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes.Crossref | GoogleScholarGoogle Scholar | 8688004PubMed |
[21] Hong, S.H. et al. (2009) Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 3, 1365–1373.
| Polymerase chain reaction primers miss half of rRNA microbial diversity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsVGru7zF&md5=63d954b58b9f30b2b37117aed7f5a37dCAS |
[22] Pinto, A.J. and Raskin, L. (2012) PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS ONE 7, e43093.
| PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1ejtb7L&md5=4c107bd9b7d4dd3732b11208bb25934eCAS | 22905208PubMed |
[23] Haas, B.J. et al. (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504.
| Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjt1aisbc%3D&md5=bc38adbbe6e29dd6df4f0f92ee79f8ebCAS | 21212162PubMed |
[24] Martin-Laurent, F. et al. (2001) DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67, 2354–2359.
| DNA extraction from soils: old bias for new microbial diversity analysis methods.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXjtlGmsLg%3D&md5=9fb876700e13990ffc91e72d6e408652CAS | 11319122PubMed |
[25] Terrat, S. et al. (2012) Molecular biomass and MetaTaxogenomic assessment of soil microbial communities as influenced by soil DNA extraction procedure. Microb. Biotechnol. 5, 135–141.
| Molecular biomass and MetaTaxogenomic assessment of soil microbial communities as influenced by soil DNA extraction procedure.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtVKmtb0%3D&md5=1be9e2d3ce62092dc3834be5d8a0484dCAS | 21989224PubMed |
[26] Lombard, N. et al. (2011) Soil-specific limitations for access and analysis of soil microbial communities by metagenomics. FEMS Microbiol. Ecol. 78, 31–49.
| Soil-specific limitations for access and analysis of soil microbial communities by metagenomics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1CitLvO&md5=559223b2bc51ec4462def3590ece9c8dCAS | 21631545PubMed |
[27] Young, J.M. et al. (2014) Limitations and recommendations for successful DNA extraction from forensic soil samples: A review. Sci. Justice 54, 238–244.
| Limitations and recommendations for successful DNA extraction from forensic soil samples: A review.Crossref | GoogleScholarGoogle Scholar | 24796953PubMed |
[28] Lee, C.K. et al. (2012) Groundtruthing next-gen sequencing for microbial ecology – biases and errors in community structure estimates from PCR amplicon pyrosequencing. PLoS ONE 7, e44224.
| Groundtruthing next-gen sequencing for microbial ecology – biases and errors in community structure estimates from PCR amplicon pyrosequencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtlCitrvK&md5=0623394bab71d8111dd20a78c83e40b2CAS | 22970184PubMed |
[29] Tedersoo, L. et al. (2010) 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 188, 291–301.
| 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXht1aqsLfK&md5=b449e3549a3e8a9d0dade7a8558375adCAS | 20636324PubMed |
[30] Gomez-Alvarez, V. et al. (2009) Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3, 1314–1317.
| Systematic artifacts in metagenomes from complex microbial communities.Crossref | GoogleScholarGoogle Scholar | 19587772PubMed |
[31] Delmont, T.O. et al. (2013) Mastering methodological pitfalls for surviving the metagenomic jungle. Bioessays 35, 744–754.
| Mastering methodological pitfalls for surviving the metagenomic jungle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtFemsr7N&md5=e8326d088bf006c6b97006688802eaf3CAS | 23757040PubMed |
Biographies
Nick Coleman gained a PhD (Microbiology) from USyd (2000), and worked as a postdoc at Tyndall Air Force Base (USA, 2000–2003) and at USyd (2003–2006), then began as a Lecturer at USyd in 2006. Nick is a member of the ASM NSW Branch Committee and the ASM2015 Canberra Local Organising Committee.
Jacob Munro completed his BSc(Adv) at USyd in 2011, graduating with Honours in Microbiology. He is currently studying for a MSc (IT-Bioinformatics) degree at UNSW. Jake’s research interests include bioinformatics, biotechnology and microbial ecology.
Deborah Rich studied a BSc(Adv) at USyd, graduating in 2014 with Honours in Microbiology. Deb is starting a PhD in the Coleman lab, studying the interactions of plants and ethylene-oxidising-bacteria, and the potential uses of these bacteria for delaying ripening and spoilage.
Simon Dingsdag did his BSc at USyd, graduating in 2010 with Honours in Microbiology. Simon is currently studying for his PhD (USyd) at the Westmead Centre for Dental Research. His research interests include microbial ecology and oral microbiology.


