Reassessment of the subspecific status of the Australian Wet Tropics yellow-bellied glider, Petaurus australis
Steven J. B. Cooper A B * , Kenny J. Travouillon C , Kristofer M. Helgen B D , Kathleen Saint A , Rupert Russell E and John Winter FA South Australian Museum, North Terrace, Adelaide, SA 5000 Australia.
B Environment Institute, School of Biological Sciences, The University of Adelaide, SA 5005 Australia.
C Western Australian Museum, Collections and Research, 49 Kew Street, Welshpool, WA 6106, Australia.
D Australian Museum Research Institute, Australian Museum, Sydney, NSW 2010, Australia.
E PO Box 63, Mt Molloy, Qld 4871, Australia.
F PO Box 1485, Atherton, Qld 4883, Australia.
Australian Mammalogy 45(2) 220-236 https://doi.org/10.1071/AM22022
Submitted: 30 June 2022 Accepted: 15 November 2022 Published: 11 January 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the Australian Mammal Society. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
The Wet Tropics (WT) population of the yellow-bellied glider Petaurus australis Shaw, 1791 in North Queensland is listed as Endangered at the state and national level, but its taxonomic classification is currently uncertain. Here we expand on previous genetic and morphological analyses of the WT population with additional samples and genetic loci to re-assess its subspecific status. Phylogenetic analyses of the mitochondrial NADH dehydrogenase subunit 4 (ND4) gene showed that the WT population formed a reciprocally monophyletic group relative to a group comprising P. australis from its remaining distribution in Australia. The genetic distinction of the WT population was further corroborated by analyses of the nuclear gene von Willebrand factor. Molecular clock analyses of combined ND2–ND4 data suggested that the WT population has been isolated from neighbouring populations in southern Queensland over multiple ice age cycles. Morphological analyses show that the WT gliders are smaller, with proportionally shortened faces, and have paler bellies compared to southern yellow-bellied gliders. We, therefore, propose that the WT population be recognised as a distinct subspecies which we herein describe. This taxonomic reassessment of P. australis has important implications for the ongoing conservation management of the WT population and yellow-bellied gliders throughout Australia.
Keywords: aspidonym, Endangered species, Evolutionarily Significant Unit, marsupial, phylogenetics, subspecies, systematics, taxonomy.
Introduction
Petaurid gliders are widespread in their distribution across native forest regions of eastern and northern Australia (and northward to New Guinea and adjacent islands), but they have suffered declines in population size due to habitat clearance for agriculture and forestry, resulting in habitat fragmentation that is reducing migration and gene flow across the landscape (Goldingay et al. 2013; Malekian et al. 2015). Their distribution was also impacted historically through Pleistocene climatic changes, which led to substructuring of populations following the replacement of wet forest by open sclerophyll woodlands (e.g. Petaurus norfolcensis (Kerr, 1792); Pavlova et al. 2010).
The yellow-bellied glider, Petaurus australis Shaw, 1791, occurs in only a single isolated population in South Australia and is patchily distributed from Victoria through to northern Queensland. It is listed as Endangered or Vulnerable in three of the four states in which it occurs and hence there is a need for conservation management programs to be implemented. Such programs, which may require translocation of animals among populations to boost genetic diversity, should be based on a sound taxonomy of the species that takes into account the genetic distinctiveness of populations and their history of connectivity (Weeks et al. 2011).
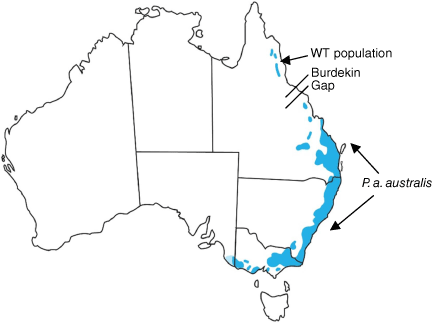
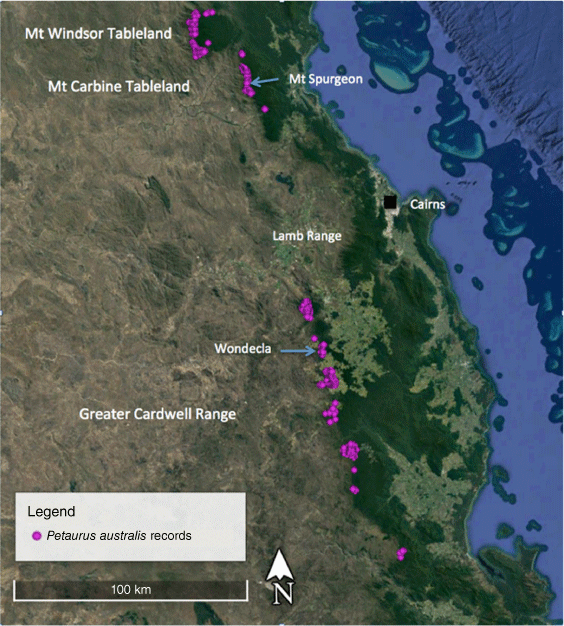
The Wet Tropics (WT) population of the yellow-bellied glider in northern Queensland, referred to as Petaurus australis unnamed subspecies, is the most northerly population of the species in Australia, and is isolated from more southerly populations on the Clarke Ranges by the dry Burdekin Gap, spanning a distance of approximately 400 km (Fig. 1; Brown et al. 2006; Van Dyck and Strahan 2008; Jackson and Groves 2015). The WT population is further subdivided into three subpopulations – on the Mt Windsor Tableland, Mt Carbine Tableland and the Greater Cardwell Range (GCR; Fig. 2) – and is restricted to a narrow band of tall eucalypt forest on the western margins of the rainforest (Winter et al. 2004; Williams 2006). The three WT subpopulations are separated by natural habitat breaks of dry eucalypt woodland not used by the gliders. The Mt Windsor Tableland and Mt Carbine Tableland populations are separated by a gap of 15 km and a saddle of 750 m in elevation of extremely dry woodland. Seventy five km separates the Mt Carbine Tableland and the GCR subpopulations, with only a narrow band of suitable habitat on the western slopes of the Lamb Range that is not known to be occupied by the gliders. The GCR sub-population is further subdivided by natural breaks in the narrow band of the glider’s natural habitat, exacerbated by recent clearing for agriculture, particularly at the northern end in the Wondecla region.
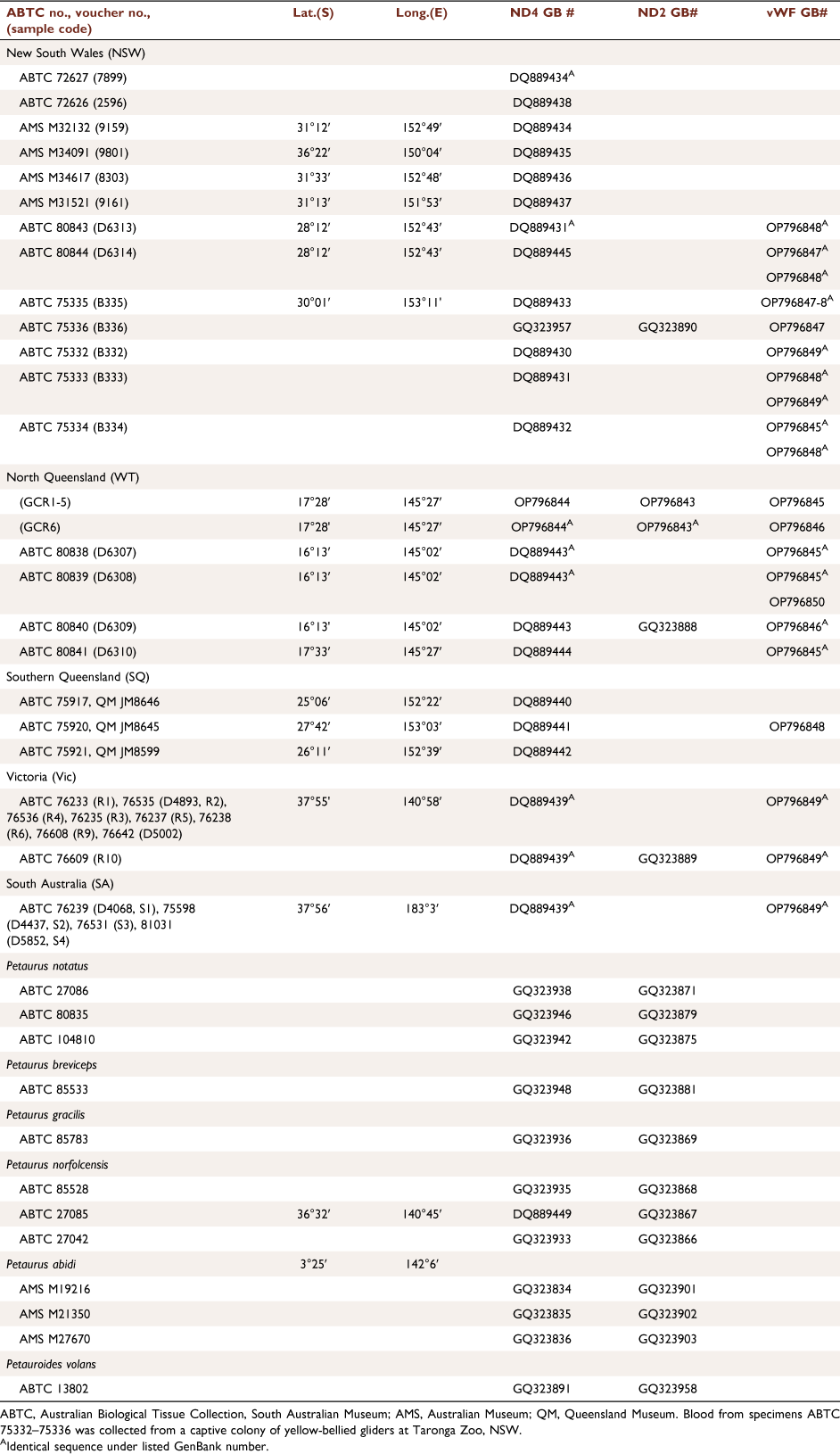
|
|
The WT population was first listed as a subspecies by Russell (1983) and allocated the subspecific name reginae Thomas, 1923. McKay (1988) also recognised P. a. reginae as a distinct subspecies comprising the WT population, and Flannery (1994) explicitly restricted the name reginae to the WT portion of the range, recognising it as a distinct WT endemic subspecies, P. a. reginae. However, this name was first applied (based largely on fur colour differences) to populations, not from the Wet Tropics, but from southern Queensland, with the type specimen coming from Gin Gin near Bundaberg (Thomas 1923), south of the Burdekin Gap (Fig. 1). Fur colour was later shown to vary with the age of individuals (Goldingay and Kavanagh 1990; Goldingay et al. 2001), darkening as the animal matures, initially casting doubt on the subspecific status of reginae and the need for the use of trinomial designations in P. australis. Nevertheless, it was proposed by Maxwell et al. (1996) that the WT population be called an ‘unnamed subspecies’ and this proposal was recognised as such by Groves (2005) and Clayton et al. (2006). Brown et al. (2006) carried out molecular (mitochondrial DNA, mtDNA) and morphological analyses of P. australis and concluded that the subspecies reginae was no longer valid, as the southern Queensland population was genetically more closely related to P. australis australis from NSW, Vic and SA compared to individuals from WT. Individuals from the latter population formed a reciprocally monophyletic group relative to a second group comprising individuals from all other populations, from southern Queensland to SA (Brown et al. 2006). Sequence data from one mtDNA locus, NADH Dehydrogenase subunit 4 (ND4) gene) from four animals, including just one sample from the GCR region, representing the largest sub-population in WT, was considered insufficient to delineate the WT population as a distinct subspecies, though the authors recommended it be treated as a distinct Evolutionarily Significant Unit (ESU) for the purposes of conservation management. The WT population is currently listed as Endangered under the Queensland Nature Conservation Act 1992 and Endangered under the Commonwealth Environment Protection and Biodiversity Conservation Act 1999, both of which recognise its subspecific status, albeit unnamed.
In the current study, we obtained additional samples from the GCR subpopulation of North Queensland to supplement those obtained by Brown et al. (2006) and carried out phylogenetic analyses of mtDNA and nuclear gene markers to further assess the subspecific status of the WT population. We also examined patterns of morphological variation in museum specimens of Petaurus australis collected throughout the range of the species. We employed criteria for defining subspecies using the concept first proposed by Avise and Ball (1990) and O’Brien and Mayr (1991) who defined subspecies as ‘groupings of populations within species that share a unique geographic range or habitat and are distinguishable from other subdivisions of the species by multiple, independent genetically based traits’ (as stated in Frankham et al. 2002, p. 617).
Methods
Tissue samples
DNA samples (n = 30) for P. australis were sourced from the South Australian Museum (from the study of Brown et al. 2006), with the exception of six new glider samples that were collected from the Wondecla area of the GCR in north Queensland WT (Table 1, Fig. 2). The latter gliders were captured at night by hand by RR when they were feeding on sap at cuts made by them on the trunks of Eucalyptus resinifera. They were transferred to a cloth bag on-site and the tip of each ear removed by JW, with a new sharp disposable scalpel blade. Tissue samples were stored in tubes of 100% ethanol. To reduce stress to the animal it was held for the minimum amount of time, no more than 5 min, before being released back onto the tree. Consequently, no body measurements were taken from the GCR population. The animals appeared little concerned by their handling and were regularly seen the following night on the same tree, showing little interest in the capturer and coming again within capture distance. They were also seen weeks later, recognised by the missing ear tips. These six animals were members of two family groups occupying adjacent territories. The tissue samples were sent to the South Australian Museum for genetic analyses. The ear biopsies were collected under ethics and scientific cover of the Threatened Species Unit of the Queensland Department of Environment and Heritage Protection (SA2013/07/432).
Genetic analyses
DNA was extracted from skin or liver tissue using the Gentra Puregene extraction kit and methods specified by the manufacturer (Gentra Systems Inc.).
Our previous molecular analyses of P. australis (Brown et al. 2006) used a ~900-bp segment of the ND4 gene, therefore we also used this marker in the current study. ND4 was amplified using the primers M245 (5′-TGA CTA CCA AAA GCT CAT GTA GAA GC-3′) and M246 (5′-TTT TAC TTG GAT TTG CAC CA-3′), each based on primers ND4 and Leu designed by Arevalo et al. (1994). We also sequenced a portion of the NADH dehydrogenase subunit 2 gene (ND2) using the primers M635 (5′-GCA CCA TTC CAC TTY TGA GT-3′) and M636 (5′-GAT TTG CGT TCG AAT GTA GCA AG-3′) (Osborne and Christidis 2001). Two nuclear genes were also sequenced: a 945 bp fragment of the vonWillebrand factor gene (vWF exon 28), using primers G807 (5′-GAC TTG GCY TTY CTS YTG GAT GG-3′) and G2526 (5′-TTG ATC TCA TCS GTR GCR GGA TTG C-3′ (Amrine-Madsen et al. 2003), and a 700 bp fragment of the ω-globin gene using primers G314 (5′-GGA ATC ATG GCA AGA AGG TG-3′) and G424 (5′-CCG GAG GTG TTY AGT GGT ATT TTC-3′) (Wheeler et al. 2001). Each of these genes have been used in previous phylogenetic studies of petaurids (Malekian et al. 2010a, 2010b).
PCR amplifications were carried out in 25 μL volumes containing 0.1U AmpliTaq Gold® polymerase (Applied Biosystems), 1 × AmpliTaq Gold Buffer, 0.20 mM dNTPs, 2.5 mM MgCl2, 0.5 μM of each primer and approximately 100 ng genomic DNA. Thermocycling conditions were: initial activation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 48–55°C for 45 s, and extension at 72°C for 60 s; and a final extension at 72°C for 3 min. PCR products were purified using Millipore MultiScreen PCR 384 Filter Plates (Millipore) and were capillary sequenced by the Australian Genome Research Facility (AGRF) using the ABI Prism Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems). Sequencing was carried out on an ABI 3700 DNA analyser and edited using SEQED ver. 1.0.3 (Applied Biosystems). Sequences were submitted to GenBank (Table 1).
Phylogenetic and haplotype network analyses
DNA sequences were edited and aligned using the Geneious alignment option within Geneious (version 9.1.2; www.geneious.com). Pairwise distances among mitochondrial haplotypes were determined using Geneious using the HKY-85 (Hasegawa et al. 1985) model of sequence evolution. Geneious was also used to construct Neighbour Joining (NJ) trees using the same model. The robustness of nodes in the NJ trees was assessed by 1000 bootstrap replicates.
The ND2 data were also phylogenetically analysed using Maximum likelihood (ML) as implemented in the program RAxML (version 7.2.8; Stamatakis et al. 2008) provided as a plugin for the program Geneious (version 9.1.2). A single model of evolution, General Time Reversible (GTR) model (Rodríguez et al. 1990) with unequal variation at sites modelled using a Gamma (G) distribution (Yang 1996) was applied to the sequence data. Robustness of branches on the tree was assessed using 500 bootstrap pseudoreplicates. Trees were visualised and rooted using the petaurids Dactylopsila trivirgata and Gymnobelideus leadbeateri as outgroups for analyses including Petaurus species or using a midpoint rooting approach in FigTree (version 1.4.2; http://tree.bio.ed.ac.uk/), for analyses of P. australis ND4 data only.
The vWF sequence data showed low levels of divergence (i.e. haplotypes differing by 1–2 nucleotide sites) within P. australis and, therefore, a haplotype network approach was used to visualise relationships among haplotypes from P. australis populations. Ambiguities in vWF sequences, representing polymorphic variation, were resolved into distinct haplotypes for each individual and an unrooted NJ network, based on HKY-85 distances (Hasegawa et al. 1985) among haplotypes, was derived using Geneious. This network was exported as a NEWICK tree and then converted into a haplotype network using the program Haploviewer (developed by G. Ewing, http://www.cibiv.at/~greg/haploviewer).
An assessment of how genetic variation was partitioned among groups of P. australis populations was determined using Analyses of Molecular Variation (AMOVA), using the program Arlequin (v. 3.5.2.2; Excoffier and Lischer 2010), with default options based on the frequency of pairwise differences (p-distances) among haplotypes in populations. This approach was used to compare different hypotheses of subspecies boundaries for both ND4 and vWF data.
Molecular clock analyses
To estimate the coalescent time of P. australis mtDNA haplotypes, molecular clock analyses were conducted on the combined ND2–ND4 sequence data using BEAST version 2.4.7 (Bouckaert et al. 2014). Exemplar sequences from Australian Petaurus species, the New Guinean P. abidi and the outgroup taxon Petauroides volans, were included in the analyses. First, second and third codon positions of each gene, were analysed as separate partitions using the program PartitionFinder (version 1.1.1; Lanfear et al. 2012) to find an appropriate partitioning scheme and models of nucleotide evolution for each partition in the BEAST analyses. The best partitioning scheme based on the Bayesian Information Criterion suggested combining first, second and third codon positions of each gene, resulting in three partitions for phylogenetic analysis: ND2–ND4 first, ND2–ND4 second, ND2–ND4 third, with the models HKY+G, HKY+I and TrN+I (Hasegawa et al. 1985; Tamura and Nei 1993; Yang 1996), respectively. The program BEAUti was used to define the site substitution models for each partition and assign molecular clock models and tree and parameter priors for different analyses. To calibrate the molecular clock a divergence time of 4.46 ± 0.1 Ma (mega annum) for the appearance of Petaurus in the fossil record was used as a minimum bound. These fossils were a mixed sample of teeth from deposits at Hamilton Victoria that were considered by Turnbull et al. (2003) to be near P. australis and P. norfolcensis. Hence, the 4.46 Ma date was placed on the common ancestral node of these two species = common ancestor of all the known extant Petaurus species (Malekian et al. 2010b). A lognormal distribution with an offset of 4.46 Ma, a mean (m) = 1.0 and a standard deviation (s) = 1.25 was used as the prior distribution for this node calibration. A birth-death tree prior, using estimated birth and death rates and uniform priors for each parameter was used in all the analyses. Molecular clock models and trees were linked across each partition. To estimate the time to most recent common ancestor (tmrca) of the P. australis root a uniform prior ranging between 0.0 and 4.46 Ma was placed on this node. Finally, two different molecular clock models were used for comparison: a strict clock model and an uncorrelated lognormal model; the prior distribution of the molecular clock rate under a strict clock model was given a Gamma distribution with alpha = 0.001 and beta = 1000; the prior distribution of the uncorrelated lognormal model used an exponential prior distribution with m = 10.0 for the mean and a Gamma prior distribution with alpha = 0.5396 and beta = 0.3819 for the standard deviation.
BEAST analyses were run for 15 million generations in multiple independent runs to test for convergence, sampling parameters and trees every 1500 generations, using the CIPRES Science Gateway (Miller et al. 2010). The program Tracer 1.6 (Rambaut et al. 2013) was used to evaluate convergence of parameter estimates following a burnin of 10%. Treeannotator (version 2.4.7) was used to generate a maximum clade credibility tree of the 15 000 trees that were sampled, using a burnin of 10% (1500 trees). Trees were visualised and initially prepared for publication using the program FigTree (version 1.4.2).
Morphological analyses
Museum specimens discussed here are deposited in the collections of the Australian Museum, Sydney (AMS), the American Museum of Natural History, New York (AMNH), the Natural History Museum, London (NHMUK), the Australian National Wildlife Collection, Canberra (ANWC), the Queensland Museum, Brisbane (QM), the Museum of Comparative Zoology at Harvard University, Cambridge, Massachusetts (MCZ), Museum Victoria, Melbourne (NMV), and the South Australian Museum (SAMA). Craniodental measurements were taken with handheld digital calipers graduated to the nearest 0.01 mm (Supplementary Table S1). Measurements and statistical comparisons are based on adult specimens as judged by direct examinations of dental and sutural maturity in skulls. Measurements taken include the greatest length of skull (GLS), condylobasal length (CBL), zygomatic width (ZYG), length of upper molars (M1-4), width of M1 (WM1), maximum width of palate (MWP), maximum breadth of braincase (BBC), height of braincase from the basioccipital floor to the apex of the braincase (HBC), nasal length (NL), nasal maximum width (NW), length of the lower incisor (length inc) and depth of the lower incisor (depth inc) (Supplementary Table S1). The dental formula of each specimen was also examined, following Archer (1984).
Body mass measurements of the southern (latitudes −35.5° to −37.5°; Victoria and southern New South Wales), central (latitudes −25° to −28°; southern Queensland) and northern populations (latitudes −17.4° to −17.9°; north Queensland’s Wet Tropics) were used to perform a box plot analysis and two-way and one-way ANOVAs, comparing each population and sexes within populations. Measurements taken on skulls were analysed using a principal component analysis (PCA). The analysis was performed both on raw measurements and on transformed measurements, after the effects of allometry were removed, using the software PAST (Hammer et al. 2001). Because several skulls were unsexed, we performed the analyses with and without defining the sex, to detect whether they helped or muddled the results. A MANOVA was also performed on each data set to identify if the means were significantly different from one another.
Specimens from the AMNH and MCZ, representing a larger sample of the northern population, did not have all measurements taken (only two) and couldn’t be used in the PCA. To include them and have a larger sample size, we produced an X–Y graph using those two measurements, using both raw and transformed measurements (allometry effects removed), and including and excluding unsexed skulls.
Results
Genetic analyses
Six animals were caught, ear biopsies taken (GCR1-6), and released, all from the Wondecla area within the GCR subpopulation in WT (Fig. 2). These six samples were PCR-amplified and sequenced for ω-globin (750 bp), ND4 (884 bp), ND2 (696 bp) and vWF (945 bp). The sequences of ω-globin from the GCR specimens were found to be identical to each other and to ω-globin sequences from P. australis from western Victoria (R9 and R10). Due to a lack of variation this marker was dropped from further analyses.
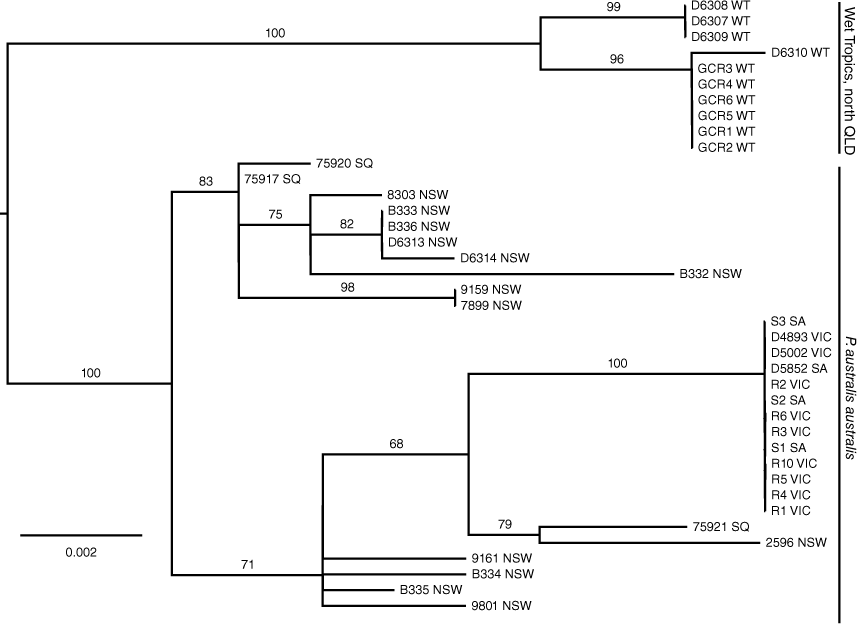
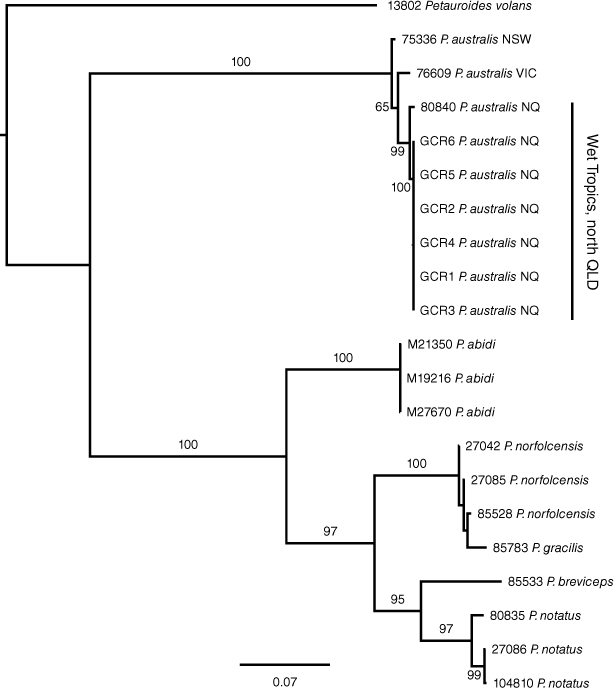
The ND2 sequences from the six GCR specimens were found to be identical, and most closely related to an ND2 haplotype from a specimen (80840) from Mt Windsor National Park, WT (p-distance = 0.077%) and 2.2–2.5% divergent from P. australis from western Victoria and NSW (76609 and 75336). Similarly, the ND4 sequences were identical for the GCR specimens, 0.6% divergent from 80840 from WT and between 1.6 and 1.7% divergent from P. australis from western Victoria and NSW (76609 and 75366) respectively. Phylogenetic analyses of individual genes, ND2 and ND4, produced identical gene trees, providing evidence that these sequences were derived from the same genetic locus and, therefore, most likely mitochondrial in origin (results not shown). Phylogenetic analyses of concatenated ND2 and ND4 data using ML showed strong support (bootstrap support (BS) = 99%) for a monophyletic group comprising all the WT samples to the exclusion of P. australis from NSW and Victoria (Fig. 3). However, the position of the root into the P. australis clade was not well supported (BS = 65), possibly due to the long branch connecting the outgroup taxa (Petauroides volans (Kerr, 1792), P. abidi, P. norfolcensis and P. breviceps Waterhouse, 1839) to the ingroup. These analyses further showed that P. australis is a sister lineage to the other species in the Petaurus genus, an arrangement that was strongly supported (BS = 100%).
|
Using the new ND4 data from WT, together with ND4 data (n = 30) from P. australis derived by Brown et al. (2006), we further analysed the phylogeographic structure evident within the species using ML analyses (Fig. 4). These analyses showed the presence of two divergent monophyletic groups of ND4 haplotypes, one comprising samples from WT and a second group comprising samples from SQ, NSW, Victoria and SA. Each group was supported by a BS value of 100%. A NJ tree was also converted into a haplotype network to provide an alternative view of the ND4 haplotype relationships, showing the presence of three distinct haplotypes in the WT population that were at least 13 mutational steps away from the most closely related haplotype from a SQ location (Fig. 5a).
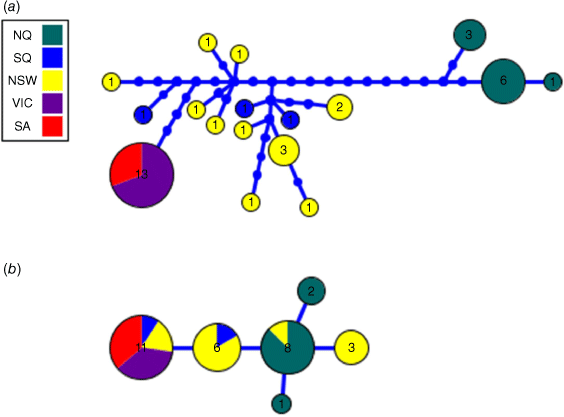
|
AMOVA analyses of the ND4 data revealed significant FST values (P = 0.000) among sub-populations within a two population structure model and that a maximum of variation among groups of 53.65% was obtained when WT was specified as a distinct population relative to a population comprising samples from SQ, NSW, Vic and SA. All other combinations of sub-populations under a two population structure model revealed lower levels for the partitioning of among group genetic variation (<29%). The optimum population structure model, however, comprised three populations: (WT), (SQ, NSW) and (Vic, SA), which explained 76% of among group genetic variation. This genetic variation among SQ-NSW and Vic-SA can largely be attributed to the presence of a single fixed haplotype in the latter population which is at the extreme western end of the distribution of P. australis.
vWF sequence analyses
A 945 bp segment of the vWF gene was sequenced from 25 specimens, with 6 specimens showing heterozygosity for alternative haplotypes. These latter sequences showed variation at a single nucleotide site, and hence could be unambiguously resolved into distinct haplotypes, giving a dataset of 31 sequences that were used to construct a haplotype network (Fig. 5b). The network shows the presence of six haplotypes of which two were found exclusively in WT specimens only, and a third haplotype (n = 8) that was found in seven WT specimens and one specimen from NSW.
AMOVA analyses of the vWF data also revealed significant FST values among sub-populations within groups for a two population structure model. For a structure comprising WT versus SQ, NSW, Vic/SA (i.e. the subspecies hypothesis), 29.5% of genetic variation was partitioned among the groups, but the optimum model of population structure comprised three populations: (WT), (SQ, NSW) and (Vic, SA), which explained 52.2% of among group genetic variation.
Molecular clock analyses of the ND2–ND4 data
The ND2–ND4 data set was further analysed using molecular clock Bayesian analyses to estimate the coalescent time of mtDNA haplotypes from the WT population relative to those from NSW and Vic. Initial analyses using the Partition Finder substitution models (HKY+G, HKY+I and TrN+I for 1st, 2nd and 3rd codon positions respectively) failed to reach convergence for the parameter of the invariant sites model despite multiple attempts to tune this parameter. Therefore, a Gamma model of rate variation (Yang 1996) was used for each of the partitions to model site specific rate variation. Estimated sample size (ESS) values for all parameters were >312, suggesting an adequate sample of the posterior distribution for each parameter had been obtained. Under a strict molecular clock model the tmrca estimate for P. australis was 169.3 thousand years (95% posterior distribution (PD) ranged from 102.3 to 248.6 thousand years). Under an uncorrelated lognormal relaxed clock model the estimate for the P. australis MRCA was 137.5 thousand years (95% PD from 76.4 to 207.6 thousand years; Supplementary Fig. S1).
Morphology
Skulls were examined for morphological differences. Skulls in the northern WT population were overall smaller with a shorter snout, compared to the southern population. The dental formula for the upper dentition (I1-3, C1, P1-3, M1-4) didn’t differ from that identified for P. australis by Archer (1984). The lower dentition was, however, different between the northern and southern populations. The southern population had the dental formula reported by Archer (1984), i1-2, p1-3, m1-4, but the northern population had variable numbers of unicuspid teeth (i2-p3), as high as four, as in the southern population, and as low as two.
We compared weights of wild-caught P. australis from across their distribution (see Fig. 6 caption for data sources) and found that they varied significantly between southern and northern populations (two-way ANOVA, F1,147 = 67.4, P < 0.0001) and between genders (F1,147 = 9.1, P = 0.003), with the southern population and males being heavier. However, the interaction between population and gender was also significant (F1,147 = 8.7, P = 0.004), indicating that the relationship between the sexes differs between northern and southern populations (Fig. 6). In the southern population, males are markedly heavier than females (mean 601 cf. 543 g, n 20 and 16), whereas in the northern population the difference is slight (mean 493.2 cf. 492.5 g, n 58 and 55) and not statistically significant (one-way ANOVA, F1,112 = 0.1, P = 0.94).
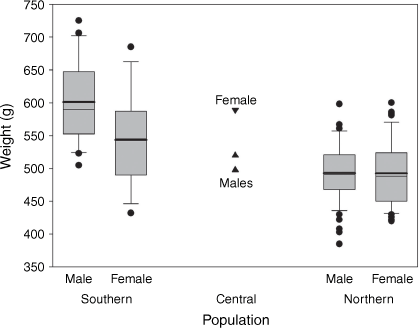
|
Morphometrics
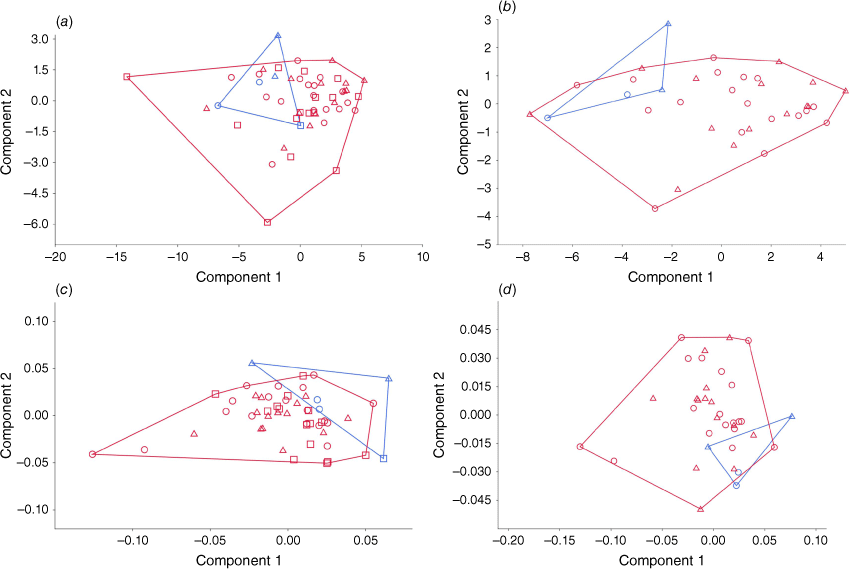
The results of the Principal Component Analysis (PCA) of cranial measurements, show a significant amount of overlap between the southern and northern populations (Fig. 7). Using all the raw measurement available (Fig. 7a), including from unsexed skulls, specimens from the northern population are within the range of the southern population. One unsexed specimen (SAMA M2237) is large and fits perfectly within the measurements of the southern population. The MANOVA found that the two populations were not significantly different (F = 1.725; P = 0.09868). If unsexed specimens are removed (Fig. 7b), the northern population overlaps less so with the southern population, occupying the top left corner of the PCA with negative component 1 values, while the majority of the southern population had positive values on component 1. The MANOVA found that the northern and southern populations have significantly different means (F = 2.997, P = 0.01148). After transforming the measurements by removing the effects of allometry (option selected in PAST), using all specimens (Fig. 7c), the northern population only overlaps slightly with the southern population, showing that the difference between the two populations is not only driven by size, but by selection in the shape of the skull. The MANOVA found a significant difference between the two populations (F = 2.915; P = 0.005693). When unsexed specimens were removed (Fig. 7d), the same results were found, corroborated by the results of the MANOVA (F = 3.222, P = 0.007707).
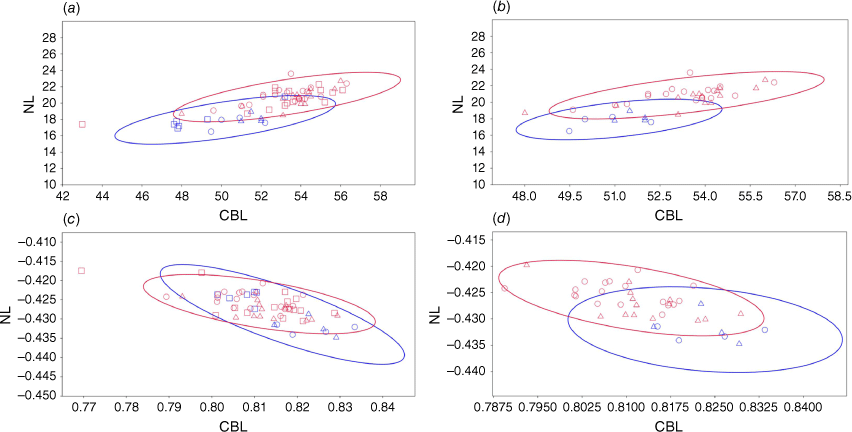
Condylobasal length (CBL) is plotted against nasal length (NL) in Fig. 8. The sample size was large enough to be able to provide 95% ellipses, with any samples outside of the ellipses considered an outlier or as being distinct. Using all specimens’ raw measurements (Fig. 8a), most of the specimens of the northern population fall outside the 95% ellipse of the southern population, suggesting that they are distinct. SAMA M2237, which was found previously to be too large to be from the northern population, is just at the edge of the northern population 95% ellipse. Two specimens from the Southern population are found outside of the 95% ellipse, SAMA M12590 (unsexed) and NMV C2396.14 (female), suggesting that they are outliers or distinct. When unsexed specimens are removed (Fig. 8b), there is much less overlap between the two 95% ellipses. NMV C2396.14 (female) is again found outside the ellipse for the southern population, as well as AMS M357 (male). When the measurements are transformed to remove the effects of allometry (Fig. 8c), the two 95% ellipses overlap each other significantly, with only a couple of specimens from the northern population outside of the southern population ellipse. SAMA M12590 (unsexed) and NMV C2396.14 (female) are found again outside of the southern population ellipse, though SAMA M12590 (unsexed) much more than NMV C2396.14 (female). When unsexed individuals are removed (Fig. 8d) the two ellipses overlap much less, with half of the northern population specimens outside of the southern population ellipse. NMV C2396.14 (female) and AMS M357 (male) remain the most distinct within the southern population, though they are within the 95% ellipse.
Discussion
Our phylogenetic and haplotype network analyses based on mtDNA and nuclear gene loci, when combined with the morphometric analyses that show that the WT specimens had a shortening of the rostrum compared to southern specimens, provide strong support for the distinctiveness of the WT population of P. australis. We further discuss these analyses below, provide additional support from ecological studies for the distinctiveness of the WT population, and propose that it should be recognised as a distinct subspecies that we describe below.
Genetics
Mitochondrial and nuclear gene data from across the range of P. australis in Australia support the presence of two major genetic lineages, one distributed in WT and a second distributed from southern Queensland to SA (Figs 3, 4). These analyses further support the conclusion from Brown et al. (2006) that the WT population represents a distinct ESU, relative to an ESU comprising all other localities in southern QLD, NSW, Vic and SA, using criteria for defining ESUs proposed by Moritz (1994). Under these criteria each ESU is reciprocally monophyletic for mtDNA loci and shows significant allele frequency variation at nuclear DNA markers.
Based on the coalescent time for the mtDNA haplotypes, the two lineages are likely to have been isolated for >130 thousand years, indicative of genetic isolation over multiple ice age cycles. Although equating population divergence dates to the coalescent time of a single genetic locus is subject to error, they nevertheless suggest that the WT lineage has been evolving on an independent evolutionary trajectory compared to southern populations over a long time period that is significant within the evolutionary history of the species. The divergence among mtDNA haplotypes between WT and southern populations of P. australis (>2.2%) is similar to that found between P. norfolcensis and P. gracilis (De Vis, 1883) (>2.0%) and greater than that found at the intra-specific level within P. norfolcensis (Pavlova et al. 2010), which is distributed over a similar range. Genetic studies of the sugar glider P. breviceps reported high levels of intra-specific variation (10.4–12.2%; Malekian et al. 2010b), which has led to the recognition of two additional species (P. notatus and P. ariel) distinct from P. breviceps (Cremona et al. 2021).
Morphology
Brown et al. (2006) concluded from their examination of morphological material over the full range of Petaurus australis that ‘morphological analysis provided little evidence for discrimination of populations, although NQ specimens were generally smaller in size than southern forms.’ Our results agree partly with Brown et al. (2006), that the northern population is generally smaller in size, but we have found that skull size was not the only factor contributing to their differentiation. Our morphometric analyses that excluded the effects of allometry showed a significant difference between the northern and southern population, meaning that the shape of the skulls was significantly different. This analysis suggests that facial shortening is being selected in the northern population, by the shortening of the nasal length, with evidence of loss of teeth in the lower jaw, with fewer teeth between the front incisors and the molars. The southern population had the dental formula reported by Archer (1984), i1-2, p1-3, m1-4, with some specimens having an extra premolar. The specimens from the WT population had either the same dental formula or had fewer unicuspid teeth between i1 and m1, with the absence of one or two of those teeth. Overall, our results support the WT population as being distinct morphologically from the southern population.
Some specimens do not follow the trend observed in other specimens from the same region. For example, SAMA M2237 from Herberton (Queensland; north-east of Wondecla, see Fig. 2) is very large and fits better with the morphology seen in the southern population. It is possible that the skin and skulls of the specimens were mixed up, and that the skull belongs to a different specimen, perhaps SAMA M2745 from Lower Fitzroy Valley, which has a much smaller skull than what would be expected from that region. NVM C2396.14 from the Otway Range in Victoria also came up as an outlier in the X–Y plot, mainly because it has the largest nasal length of any specimen. All other measurements are within the range of the population, and therefore this specimen isn’t seen here as being distinct morphologically, but rather an outlier in the dataset. SAMA M12590 is, however, a real mystery. It is unsexed and from an unknown location. The comments associated with the specimen say ‘Old museum stock’, and it was previously identified as Petaurus breviceps. Our results suggest that it is so distinct from Petaurus australis, that this specimen probably represents another taxon. Finally, AMS M357 is a male from Orange (NSW), which is very small in size and has a white belly. Both features would suggest that it is distinct from the rest of the southern population specimens and should warrant genetic assessment in the future.
Additional characteristics of the WT population
Fur colour
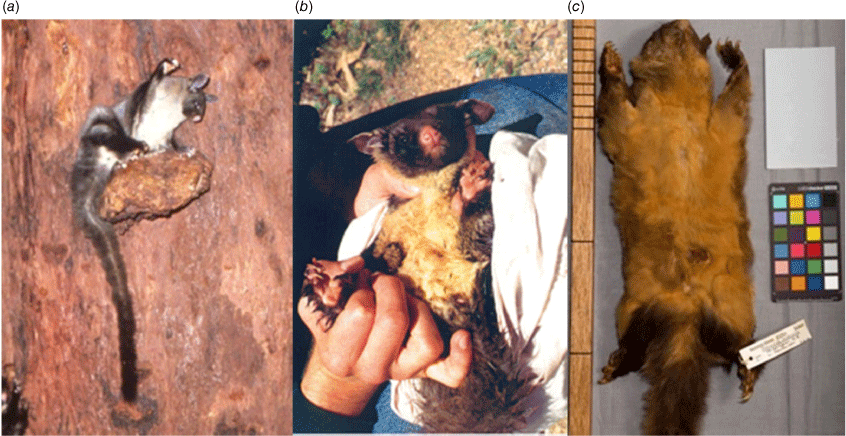
General observations by two of the authors (R. R. and J. W.) of the gliders in the most northerly population isolates in the GCR, the Mt Carbine Tableland and Mt Windsor Tableland subpopulations, noted that the gliders have paler belly fur than the rich yellow common in more southerly populations (Russell 1984) (Fig. 9a). If this is the case, it could be further evidence of a break in gene flow, providing additional support for the taxonomic distinction of the WT population. However, fur colour is often a difficult character on which to base taxonomic separation of populations as it can vary within a population and if based on museum specimens it is subject to change owing to foxing (Wagstaffe and Williamson 1947). Goldingay et al. (2001) disputed the observation of the white belly fur in WT populations as they found in their Nitchaga study population, in the central section of the GCR, that 74% had yellow or dark yellow belly fur (Fig. 9b), though they did not compare the fur colour intensity of Nitchaga animals with those from southern Australia. In addition, the type specimen we are using from Mt Spurgeon, Mt Carbine Tableland subpopulation, collected in 1937, has strong yellow colouration, even allowing for the colour becoming darker on the specimen owing to foxing (Fig. 9c). It is from the same population which RR and JW have participated in seven biennial population surveys, with missing years, since 1997 without noting strong yellow belly colour of the adult gliders. Also, the first two specimens ever collected from the Wet Tropics region, in the Herberton area in 1912, were described by Finlayson (1934) as ‘larger, much darker and more richly yellow’ than ‘three from Rocky Waterhole, Serpentine Creek, about 28 miles north-east of Rockhampton.’ The Herberton area is the locality of Russell’s intensive behavioural study of the gliders (Russell 1984) and he never noted gliders with richly yellow bellies. This raises the question: is the Wet Tropics population trending towards an increasingly higher proportion of individuals with pale belly fur? Is it a case of genetic drift (Messer et al. 2016) occurring within relatively small population fragments of the Wet Tropics gliders? It is beyond the scope of our study to determine whether this is occurring, but, nevertheless, the white belly fur is a prominent feature of WT glider populations, further supporting the suggestion that they are on an independent evolutionary trajectory, compared to populations in the south.
Socioecology
The WT population of P. australis also exhibits differences in its socioecology from southern populations. These differences are not necessarily indicative of taxonomic differences, but likely reflect different responses to their environment. WT gliders form groups of between two and six individuals (Russell 1984; Goldingay et al. 2001). The social structure (e.g. genetic relatedness of individuals) of these groups is currently unknown. The structure of the WT groups varied, usually containing an adult male and female, but frequently with two adult females as part of the group (Russell 1984; Goldingay et al. 2001). In contrast, analyses of social structure in P. australis populations from Victoria have provided evidence for social and genetic monogamy, with single males and females sharing their home ranges and dens, and with social partners shown, using microsatellite DNA analyses, to be the true parents of juveniles in the study population (Brown et al. 2007). These differences in social behaviour may have resulted from variation in food resource abundance and productivity at the different localities (Goldingay 1992; Goldingay et al. 2001). For example, the availability of nectar and pollen food resources throughout the year may promote larger group sizes and a possible polygynous mating system (Goldingay 1992; Goldingay et al. 2001; Brown et al. 2007). Further research of social behaviour of the WT population and other populations of P. australis in eastern Australia is required, but taken overall, current observations suggest that the behavioural ecology of the WT gliders is likely to be distinct from that in southern populations.
The WT population of P. australis is unusual in that the gliders tap only one species of tree for its sap, Eucalyptus resinifera (Russell 1984; Quin et al. 1996). The only other population that targets only one species, Eucalyptus viminalis, is at the other extremity of the glider’s range, Rennick Forest in Victoria on the border with South Australia (Carthew et al. 1999). Carthew et al. (1999) point out that the restriction to one tree in Rennick Forest may be attributed to the paucity of eucalypt species present, just two with a third present in some locations. This contrasts with the greater range of species available to the WT population, both within their tall eucalypt forest habitat and adjoining rainforest (Quin et al. 1996). Tree species readily available within the WT glider habitat are E. tereticornis, E. grandis, E. moluccana and Corymbia intermedia, all of which have been recorded as tapped by the gliders in southern populations (Mackowski 1988; Eyre and Goldingay 2005). Although Eyre and Goldingay (2005) state that E. tereticornis is not a favoured tree, it is tapped for its sap by the most westerly population in Queensland in the Carnarvon Ranges together with grey gum and spotted gum (T. Eyre pers. comm. October 2018). In all other localities at least two species of tree are tapped for their sap by the gliders (Henry and Craig 1984; Craig 1985; Goldingay 1992; Goldingay and Kavanagh 1993; Eyre and Goldingay 2003). In addition, two species of tree, E. resinifera and E. moluccana, are tapped in the population immediately to the south of the WT population, at Crediton, west of Mackay (J. Winter pers. obs. 1989). There is one example in the Wet Tropics of an E. grandis tapped by the gliders. It was a young coppice trunk arising from a stump and heavily incised by the gliders in the Mt Carbine Tableland sub-population (R. Russell pers. obs. 1990s). It is unclear why the gliders did not use the readily available E. grandis more often within their range.
Justification of taxonomic status
A definition of a subspecies was proposed by Lidicker (1962). He states that ‘a subspecies is a relatively homogeneous and genetically distinct portion of a species which represents a separately evolving, or recently evolved lineage, with its own evolutionary tendencies, inhabits a definite geographic area, is usually at least partially isolated, and may intergrade gradually, although over a fairly narrow zone, with adjacent subspecies’ (see Owen 1989). Similar definitions were proposed by Avise and Ball (1990) and O’Brien and Mayr (1991), which further emphasised that subspecies should show divergence for multiple independent genetically based traits.
The use of genetic criteria to define taxonomic units, particularly at the species level, has been proposed by multiple authors (e.g. Halt et al. 2009; Cook et al. 2010; Harvey et al. 2015; Westerman et al. 2016). Subspecies, however, are usually defined based on morphological traits, with genetic data used to define alternative conservation units such as ESUs where clear genetic criteria for their recognition have been proposed (e.g. Moritz 1994). However, we concur with the view of Aplin et al. (2015) that the use of ESUs is largely informal and this concept has often not been recognised in legislation for their conservation protection (e.g. the federal Department of Environment in Australia), in contrast to subspecies. The Moritz (1994) criteria for defining ESUs – reciprocal monophyly of mtDNA, reflecting long term population isolation, and allelic/haplotype frequency variation for nuclear gene markers – in addition to consideration of other criteria (geographic isolation or presence of narrow hybrid zones, ecological differences, morphological variation) provides a sound basis for formally defining subspecies. Overall, our study fulfils each of these criteria, with the WT population representing a distinct ESU, with long-term genetic and geographic isolation. It shows distinct morphological differences (shortening of the rostrum) compared to southern populations, smaller size, a trend towards a white belly colour and some ecological differences associated with its habitat and behaviour. We consider that this degree of divergence at multiple independent genetically based traits warrants its recognition as a distinct subspecies.
Since no reliable scientific name has previously been applied to P. australis from northern Queensland (McKay 1988), we here describe the WT population as a new subspecies.
Systematics
Following the recommendations of Kaiser et al. (2013) and Jackson et al. (2022) and the position adopted by Taxonomy Australia (2021, downloaded from www.taxonomyaustralia.org.au/codes-of-conduct) we consider previous names proposed for yellow-bellied glider (Wet Tropics) to be unscientific and unavailable.
Order DIPROTODONTIA Owen, 1877
Superfamily PETAUROIDEA Bonaparte, 1832
Family PETAURIDAE Bonaparte, 1832
Petaurus Shaw, 1791
Petaurus australis Shaw, 1791
Recommended common name
Northern yellow-bellied glider.
Holotype and type locality
QM J6352, adult female, study skin (figured by Van Dyck 1997) and skull, from Mount Spurgeon, Queensland, collected 8 February 1938 by Gabriele Neuhäuser (see Mather (1986) and Van Dyck (1997) for biographical notes on Neuhäuser, and context of collection). The type locality is located today within the boundaries of Mount Spurgeon National Park.
Referred specimens
There are additional specimens at AMNH (AMNH 107263–107268, collected by G. Neuhäuser in November and December 1937) and at MCZ (MCZ 29164–29168, collected by P. J. Darlington in July 1932) that were collected from Mount Spurgeon, the type locality (Tate 1952).
Six individuals sampled and released from the Wondecla area of the GCR, in addition to ABTC80841 from Brown et al. (2006) from the same region, which were included in our genetic comparisons, are referred to this new subspecies. In addition, ABTC80838–80840 from Brown et al. (2006) from Mt Windsor Tableland are referred to this new subspecies. DNA for these samples is available from the SA Museum DNA archive.
Three specimens in the Queensland Museum from Nitchaga Creek on the Atherton Tableland, Queensland, are also referred to this subspecies: QM JM8746, adult male, skull and postcranial skeleton, from Nichaga Creek, Atherton Tableland, collected 16 July 1991 by D. Quinn; QM JM8747, adult male, skull and postcranial skeleton, from Nitchaga Creek, Atherton Tableland, collected 29 April 1991 by D. Quin; QM JM8503, adult female, skull and partial postcranial skeleton, from Nitchaga Creek, Atherton Tableland, collected 10 October 1990 by D. Quin.
Two skins at SAMA are referred to this subspecies: SAMA M2236, adult skin (without any accompanying skull), and SAMA M2237, adult skin (with an accompanying skull, which may be mismatched, see above), both from Herberton, Queensland, collected by S. Grant in 1912. Both skins had their skulls removed from their study skins, presumably (based on information from the old specimen register) after arriving at the museum.
Distribution
This subspecies is restricted to very tall eucalypt forest on the western edge of rainforest in the Wet Tropics Bioregion of north Queensland at upland altitudes above 600–700 m elevation, on tableland or plateau geomorphology. It occurs from the Herbert River Gorge to the Mount Windsor Tableland and is highly likely only found north of Burdekin Gap (Fig. 2). The subspecies Petaurus australis australis is only found south of Burdekin Gap.
Diagnosis
Petaurus australis brevirostrum differs from P. a. australis in averaging smaller in skull length, and in having a proportionally shortened facial region of the skull, as reflected in its especially short nasals (Supplementary Table S1); this shortening of the face is also reflected in some specimens by having the number of lower unicuspid teeth (between i1 and m1) sometimes reduced to 2 or 3 on each side, compared to the 4 unicuspids (i2, p1-3) on each side of the lower jaw characterising P. a. australis. The two subspecies are similar in external appearance, but P. a. brevirostrum averages smaller in overall body size (Fig. 6) and has belly fur that is less distinctly yellow, often appearing white overall, compared to P. a. australis (Fig. 9).
Etymology
The name brevirostrum means short snout and is derived from Latin ‘brevis’ for short and ‘rostrum’ meaning beak or snout. The name brevirostrum refers to the shortened rostrum of gliders from the Wet Tropics. The subspecific name is used as a noun in apposition.
Supplementary material
Supplementary material is available online.
Data availability
All sequence data are available on GenBank (see Table 1 for accession codes) and morphometrics data are provided as supplementary information.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This research did not receive any specific funding, with genetic analyses funded by SJBC’s discretionary research funds in the SA Museum and morphological examinations in museums funded by KMH’s discretionary funds at the University of Adelaide.
Acknowledgements
We wish to acknowledge Rachel Barr, Steve Murphy, David Armbrust and Diana Armbrust who helped with the collection of the specimens, and Vera Bass on whose property two of the specimens were collected. We thank David Stemmer (SAMA), Kevin Rowe and Karen Roberts (NMV), Sandy Ingleby (AMS), Steve Van Dyck and Heather Janetzki (QM), Roberto Portela Miguez (NHMUK), Mark Omura (MCZ) and Robert Voss and Neil Duncan (AMNH) for access to specimens in museum collections and Dr John Sheridan for the image of the type specimen. We thank Sue Carthew for initiating our research on yellow-bellied gliders, Meredeth Brown for past genetic analyses, Ross Goldingay for discussions on coat colour morphology and for providing a distribution map for Fig. 1 and Don Franklin for analysis of weights.
References
Amrine-Madsen, H., Scally, M., Westerman, M., Stanhope, M. J., Krajewski, C., and Springer, M. S. (2003). Nuclear gene sequences provide evidence for the monophyly of australidelphian marsupials. Molecular Phylogenetics and Evolution 28, 186–196.| Nuclear gene sequences provide evidence for the monophyly of australidelphian marsupials.Crossref | GoogleScholarGoogle Scholar |
Aplin, K. P., Rhind, S. G., Ten Have, J., and Chesser, R. T. (2015). Taxonomic revision of Phascogale tapoatafa (Meyer, 1793) (Dasyuridae; Marsupialia), including descriptions of two new subspecies and confirmation of P. pirata Thomas, 1904 as a ‘Top End’ endemic. Zootaxa 4055, 1–73.
| Taxonomic revision of Phascogale tapoatafa (Meyer, 1793) (Dasyuridae; Marsupialia), including descriptions of two new subspecies and confirmation of P. pirata Thomas, 1904 as a ‘Top End’ endemic.Crossref | GoogleScholarGoogle Scholar |
Archer, M. (1984). The Australian marsupial radiation. In ‘Vertebrate Zoogeography and Evolution in Australasia’. (Eds M. Archer, G. Clayton.) pp. 633–708. (Hesperian Press: Carlisle, Western Australia.)
Arevalo, E., Davis, S. K., and Sites, J. W. (1994). Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in central Mexico. Systematic Biology 43, 387–418.
| Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in central Mexico.Crossref | GoogleScholarGoogle Scholar |
Avise, J. C., and Ball, R. M. J. (1990). Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Survey of Evolutionary Biology 7, 45–67.
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C.-H., Xie, D., Suchard, M. A., Rambaut, A., and Drummond, A. J. (2014). BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10, e1003537.
| BEAST 2: A software platform for Bayesian evolutionary analysis.Crossref | GoogleScholarGoogle Scholar |
Brown, M., Cooksley, H., Carthew, S. M., and Cooper, S. J. B. (2006). Conservation units and phylogeographic structure of an arboreal marsupial, the yellow-bellied glider (Petaurus australis). Australian Journal of Zoology 54, 305–317.
| Conservation units and phylogeographic structure of an arboreal marsupial, the yellow-bellied glider (Petaurus australis).Crossref | GoogleScholarGoogle Scholar |
Brown, M., Carthew, S. M., and Cooper, S. J. B. (2007). Monogamy in an Australian arboreal marsupial, the yellow-bellied glider (Petaurus australis). Australian Journal of Zoology 55, 185–195.
| Monogamy in an Australian arboreal marsupial, the yellow-bellied glider (Petaurus australis).Crossref | GoogleScholarGoogle Scholar |
Carthew, S. M., Goldingay, R. L., and Funnell, D. L. (1999). Feeding behaviour of the yellow-bellied glider (Petaurus Australis) at the Western Edge of Its Range. Wildlife Research 26, 199–208.
| Feeding behaviour of the yellow-bellied glider (Petaurus Australis) at the Western Edge of Its Range.Crossref | GoogleScholarGoogle Scholar |
Clayton, M., Wombey, J. C., Mason, I. J., Chesser, R. T., and Wells, A. (2006). ‘CSIRO list of Australian vertebrates: A reference with conservation status’, 2nd edn. (CSIRO Publishing: Melbourne.)
Cook, L. G., Edwards, R. D., Crisp, M. D., and Hardy, N. B. (2010). Need morphology always be required for new species descriptions? Invertebrate Systematics 24, 322–326.
| Need morphology always be required for new species descriptions?Crossref | GoogleScholarGoogle Scholar |
Craig, S. A. (1985). Social organization, reproduction and feeding behaviour of a population of yellow-bellied gliders, Petaurus australis (Marsupialia: Petauridae). Wildlife Research 12, 1–18.
| Social organization, reproduction and feeding behaviour of a population of yellow-bellied gliders, Petaurus australis (Marsupialia: Petauridae).Crossref | GoogleScholarGoogle Scholar |
Cremona, T., Baker, A. M., Cooper, S. J. B., Montague-Drake, R., Stobo-Wilson, A. M., and Carthew, S. M. (2021). Integrative taxonomic investigation of Petaurus breviceps (Marsupialia: Petauridae) reveals three distinct species. Zoological Journal of the Linnean Society 191, 503–527.
| Integrative taxonomic investigation of Petaurus breviceps (Marsupialia: Petauridae) reveals three distinct species.Crossref | GoogleScholarGoogle Scholar |
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10, 564–567.
| Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows.Crossref | GoogleScholarGoogle Scholar |
Eyre, T. J., and Goldingay, R. L. (2003). Use of sap trees by the yellow-bellied glider near Maryborough in south-east Queensland. Wildlife Research 30, 229–236.
| Use of sap trees by the yellow-bellied glider near Maryborough in south-east Queensland.Crossref | GoogleScholarGoogle Scholar |
Eyre, T. J., and Goldingay, R. L. (2005). Characteristics of sap trees used by yellow-bellied gliders in southern Queensland. Wildlife Research 32, 23–35.
| Characteristics of sap trees used by yellow-bellied gliders in southern Queensland.Crossref | GoogleScholarGoogle Scholar |
Finlayson, H. H. (1934). On mammals from the Dawson and Fitzroy valleys: central coastal Queensland. Part II. Transactions and Proceedings of the Royal Society of South Australia 58, 218–231.
Flannery, T. F. (1994). ‘Possums of the world: A monograph of the Phalangeroidea.’ (Geo 635 Productions: Sydney.)
Frankham, R., Ballou, J. D., and Briscoe, D. A. (2002). ‘Introduction to conservation genetics.’ (Cambridge University Press: Cambridge, UK.)
Goldingay, R. L. (1992). Socioecology of the yellow-bellied glider (Petaurus australis) in a coastal forest. Australian Journal of Zoology 40, 267–278.
| Socioecology of the yellow-bellied glider (Petaurus australis) in a coastal forest.Crossref | GoogleScholarGoogle Scholar |
Goldingay, R. L. (2008). Yellow-bellied glider Petaurus australis Shaw, 1791. In ‘The mammals of Australia’, 3rd edn. (Eds S. Van Dyck, R. Strahan.) pp. 228–230. (Reed New Holland: Sydney.)
Goldingay, R. L., and Kavanagh, R. P. (1990). Socioecology of the yellow-bellied glider (Petaurus australis) at Waratah Creek, New South Wales. Australian Journal of Zoology 38, 327–341.
| Socioecology of the yellow-bellied glider (Petaurus australis) at Waratah Creek, New South Wales.Crossref | GoogleScholarGoogle Scholar |
Goldingay, R. L., and Kavanagh, R. P. (1993). Home-range estimates and habitat of the yellow-bellied glider (Petaurus australis) at Waratah Creek, New South Wales. Wildlife Research 20, 387–403.
| Home-range estimates and habitat of the yellow-bellied glider (Petaurus australis) at Waratah Creek, New South Wales.Crossref | GoogleScholarGoogle Scholar |
Goldingay, R. L., Quin, D. G., and Churchill, S. (2001). Spatial variability in the social organisation of the yellow-bellied glider (Petaurus australis) near Ravenshoe, north Queensland. Australian Journal of Zoology 49, 397–409.
| Spatial variability in the social organisation of the yellow-bellied glider (Petaurus australis) near Ravenshoe, north Queensland.Crossref | GoogleScholarGoogle Scholar |
Goldingay, R. L., Harrisson, K. A., Taylor, A. C., Ball, T. M., Sharpe, D. J., and Taylor, B. D. (2013). Fine-scale genetic response to landscape change in a gliding mammal. PLoS One 8, e80383.
| Fine-scale genetic response to landscape change in a gliding mammal.Crossref | GoogleScholarGoogle Scholar |
Groves, C. (2005). Order Diprotodontia: Petauridae. In ‘Mammal species of the world: A taxonomic and geographic reference’. (Eds D. E. Wilson, D. M. Reeder.) pp. 54–55. (Johns Hopkins University Press: Baltimore.)
Halt, M. N., Kupriyanova, E. K., Cooper, S. J. B., and Rouse, G. W. (2009). Naming species with no morphological indicators: species status of Galeolaria caespitosa (Annelida : Serpulidae) inferred from nuclear and mitochondrial gene sequences and morphology. Invertebrate Systematics 23, 205–222.
| Naming species with no morphological indicators: species status of Galeolaria caespitosa (Annelida : Serpulidae) inferred from nuclear and mitochondrial gene sequences and morphology.Crossref | GoogleScholarGoogle Scholar |
Hammer, Ø., Harper, D. A. T., and Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4, 1–9.
Harvey, M. S., Main, B. Y., Rix, M. G., and Cooper, S. J. B. (2015). Refugia within refugia: in situ speciation and conservation of threatened Bertmainius (Araneae: Migidae), a new genus of relictual trapdoor spiders endemic to the mesic zone of south-western Australia. Invertebrate Systematics 29, 511–553.
| Refugia within refugia: in situ speciation and conservation of threatened Bertmainius (Araneae: Migidae), a new genus of relictual trapdoor spiders endemic to the mesic zone of south-western Australia.Crossref | GoogleScholarGoogle Scholar |
Hasegawa, M., Kishino, H., and Yano, T. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution 22, 160–174.
| Dating of the human-ape splitting by a molecular clock of mitochondrial DNA.Crossref | GoogleScholarGoogle Scholar |
Henry, S. H. (1985). The diet and socioecology of gliding possums in southern Victoria. PhD thesis, Monash University, Melbourne.
Henry, S. R., and Craig, S. A. (1984). Diet, ranging behaviour and social organization of the yellow-bellied glider (Petaurus australis Shaw) in Victoria. In ‘Possums and Gliders’. (Eds A. P. Smith, I. D. Hume.) pp. 331–341. (Australian Mammal Society: Sydney.)
Jackson, S., and Groves, C. (2015). ‘Taxonomy of Australian mammals.’ (CSIRO Publishing: Melbourne.)
Jackson, S. M., Baker, A. M., Eldridge, M. D. B., Fisher, D. O., Frankham, G. J., Lavery, T. H., MacDonald, A. J., Menkhorst, P. W., Phillips, M. J., Potter, S., Rowe, K. C., Travouillon, K. J., and Umbrello, L. S. (2022). The importance of appropriate taxonomy in Australian mammalogy. Australian Mammalogy , .
| The importance of appropriate taxonomy in Australian mammalogy.Crossref | GoogleScholarGoogle Scholar |
Kaiser, H., Crother, B. I., Kelly, C. M. R., Luiselli, L., O’Shea, M., Ota, H., Passos, P., Schleip, W. D., and Wüster, W. (2013). Best practices: In the 21st century, taxonomic decisions in herpetology are acceptable only when supported by a body of evidence and published via peer-review. Herpetological Review 44, 8–23.
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29, 1695–1701.
| PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar |
Lidicker, W. Z. (1962). The nature of subspecies boundaries in a desert rodent and its implications for subspecies taxonomy. Systematic Zoology 11, 160–171.
| The nature of subspecies boundaries in a desert rodent and its implications for subspecies taxonomy.Crossref | GoogleScholarGoogle Scholar |
Mackowski, C. M. (1988). Characteristics of eucalypts incised for sap by the yellow-bellied glider, Petaurus australis Shaw (Marsupialia: Petauridae), in Northeastern New South Wales. Australian Mammalogy 11, 5–13.
| Characteristics of eucalypts incised for sap by the yellow-bellied glider, Petaurus australis Shaw (Marsupialia: Petauridae), in Northeastern New South Wales.Crossref | GoogleScholarGoogle Scholar |
Malekian, M., Cooper, S. J. B., and Carthew, S. M. (2010a). Phylogeography of the Australian sugar glider (Petaurus breviceps): evidence for a new divergent lineage in eastern Australia. Australian Journal of Zoology 58, 165–181.
| Phylogeography of the Australian sugar glider (Petaurus breviceps): evidence for a new divergent lineage in eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Malekian, M., Cooper, S. J. B., Norman, J. A., Christidis, L., and Carthew, S. M. (2010b). Molecular systematics and evolutionary origins of the genus Petaurus (Marsupialia: Petauridae) in Australia and New Guinea. Molecular Phylogenetics and Evolution 54, 122–135.
| Molecular systematics and evolutionary origins of the genus Petaurus (Marsupialia: Petauridae) in Australia and New Guinea.Crossref | GoogleScholarGoogle Scholar |
Malekian, M., Cooper, S. J. B., Saint, K. M., Lancaster, M. L., Taylor, A. C., and Carthew, S. M. (2015). Effects of landscape matrix on population connectivity of an arboreal mammal, Petaurus breviceps. Ecology and Evolution 5, 3939–3953.
| Effects of landscape matrix on population connectivity of an arboreal mammal, Petaurus breviceps.Crossref | GoogleScholarGoogle Scholar |
Mather, P. (1986). ‘A time for a museum: The history of the Queensland Museum 1862-1986.’ (Queensland Museum: Brisbane.)
Maxwell, S., Burbidge, A. A., and Morris, K. (1996). ‘The 1996 Action Plan for Australian Marsupials and Monotremes. Endangered Species Program, Project No. 500.’ (ANCA: Canberra.)
McKay, G. M. (1988). Family Petauridae. In ‘Zoological Catalogue of Australia, 5, Mammalia’. (Ed. D. W. Walton.) pp. 87–97. (Australian Government Publishing Service: Canberra.)
Messer, P. W., Ellner, S. P., and Hairston, N. G. (2016). Can population genetics adapt to rapid evolution? Trends in Genetics 32, 408–418.
| Can population genetics adapt to rapid evolution?Crossref | GoogleScholarGoogle Scholar |
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In ‘Proceedings of the Gateway Computing Environments Workshop (GCE), 2010, New Orleans, Louisiana’. pp. 1–8. (Institute of Electrical and Electronics Engineers (IEEE): Louisiana.)
Moritz, C. (1994). Defining ‘evolutionarily significant units’ for conservation. Trends in Ecology and Evolution 9, 373–375.
| Defining ‘evolutionarily significant units’ for conservation.Crossref | GoogleScholarGoogle Scholar |
O’Brien, S. J., and Mayr, E. (1991). Bureaucratic mischief: recognizing endangered species and subspecies. Science 251, 1187–1188.
| Bureaucratic mischief: recognizing endangered species and subspecies.Crossref | GoogleScholarGoogle Scholar |
Osborne, M. J., and Christidis, L. (2001). Molecular phylogenetics of Australo-Papuan possums and gliders (Family Petauridae). Molecular Phylogenetics and Evolution 20, 211–224.
| Molecular phylogenetics of Australo-Papuan possums and gliders (Family Petauridae).Crossref | GoogleScholarGoogle Scholar |
Owen, J. G. (1989). Population and geographic variation of Peromyscus leucopus in relation to climatic factors. Journal of Mammalogy 70, 98–109.
| Population and geographic variation of Peromyscus leucopus in relation to climatic factors.Crossref | GoogleScholarGoogle Scholar |
Pavlova, A., Walker, F. M., van der Ree, R., Cesarinia, S., and Taylor, A. C. (2010). Threatened populations of the Australian squirrel glider (Petaurus norfolcensis) show evidence of evolutionary distinctiveness on a Late Pleistocene timescale. Conservation Genetics 11, 2393–2407.
| Threatened populations of the Australian squirrel glider (Petaurus norfolcensis) show evidence of evolutionary distinctiveness on a Late Pleistocene timescale.Crossref | GoogleScholarGoogle Scholar |
Quin, D. G., Smith, A. P., and Norton, T. W. (1996). Eco-geographic variation in size and sexual dimorphism in sugar gliders and squirrel gliders (Marsupialia: Petauridae). Australian Journal of Zoology 44, 19–45.
| Eco-geographic variation in size and sexual dimorphism in sugar gliders and squirrel gliders (Marsupialia: Petauridae).Crossref | GoogleScholarGoogle Scholar |
Rambaut, A., Suchard, M., and Drummond, A. J. (2013). Tracer v1.6. Available at http://tree.bio.ed.ac.uk/software/tracer/
Rodríguez, F., Oliver, J. L., Marín, A., and Medina, J. R. (1990). The general stochastic model of nucleotide substitution. Journal of Theoretical Biology 142, 485–501.
| The general stochastic model of nucleotide substitution.Crossref | GoogleScholarGoogle Scholar |
Russell, R. (1983). Yellow-bellied glider Petaurus australis. In ‘The Australian Museum Complete Book of Australian Mammals’. (Ed. R. Strahn.) p. 136. (Angus and Robertson Publishers: Sydney.)
Russell, R. (1984). Social behaviour of the yellow-bellied glider, Petaurus australis reginae in north Queensland. In ‘Possums and Gliders’. (Eds A. P. Smith, I. D. Hume.) pp. 343–353. (Australian Mammal Society: Sydney.)
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57, 758–771.
| A rapid bootstrap algorithm for the RAxML Web servers.Crossref | GoogleScholarGoogle Scholar |
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10, 512–526.
| Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees.Crossref | GoogleScholarGoogle Scholar |
Tate, G. H. H. (1952). Mammals of Cape York Peninsula, with notes on the occurrence of rain forest in Queensland. Bulletin of the American Museum of Natural History 98, 563–616.
Thomas, O. (1923). XXVI.—On some Queensland Phalangeridæ. Annals and Magazine of Natural History 11, 246–250.
| XXVI.—On some Queensland Phalangeridæ.Crossref | GoogleScholarGoogle Scholar |
Turnbull, W. D., Lundelius Jr., E. L., and Archer, M. (2003). Dasyurids, perameloids, phalangeroids, and vombatoids from the early Pliocene Hamilton fauna, Victoria, Australia. Bulletin of the American Museum of Natural History 279, 513–540.
| Dasyurids, perameloids, phalangeroids, and vombatoids from the early Pliocene Hamilton fauna, Victoria, Australia.Crossref | GoogleScholarGoogle Scholar |
Van Dyck, S. (1997). Gabriele Neuhäuser. In ‘Brilliant Careers: Women Collectors and Illustrators in Queensland’. (Ed. J. McKay.) pp. 50–52. (Queensland Museum: Brisbane.)
Van Dyck, S., and Strahan, R. (2008). ‘Mammals of Australia.’ (Reed New Holland: Sydney.)
Wagstaffe, R., and Williamson, K. (1947). Cabinet colour-changes in bird-skins and their bearing on racial segregation. British Birds 40, 322–325.
Weeks, A. R., Sgro, C. M., Young, A. G., Frankham, R., Mitchell, N. J., Miller, K. A., Byrne, M., Coates, D. J., Eldridge, M. D. B., Sunnucks, P., Breed, M. F., James, E. A., and Hoffmann, A. A. (2011). Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evolutionary Applications 4, 709–25.
| Assessing the benefits and risks of translocations in changing environments: a genetic perspective.Crossref | GoogleScholarGoogle Scholar |
Westerman, M., Blacket, M. J., Hintz, A., Armstrong, K., Woolley, P. A., and Krajewski, C. (2016). A plethora of planigales: genetic variability and cryptic species in a genus of dasyurid marsupials from northern Australia. Australian Journal of Zoology 64, 303–311.
| A plethora of planigales: genetic variability and cryptic species in a genus of dasyurid marsupials from northern Australia.Crossref | GoogleScholarGoogle Scholar |
Wheeler, D., Hope, R. M., Cooper, S. J. B., Dolman, G., Webb, G. C., Bottema, C. D. K., Gooley, A. A., Goodman, M., and Holland, R. A. B. (2001). An orphaned mammalian β-globin gene of ancient evolutionary origin. Proceedings of the National Academy of Sciences 98, 1101–1106.
| An orphaned mammalian β-globin gene of ancient evolutionary origin.Crossref | GoogleScholarGoogle Scholar |
Williams, S. E. (2006). ‘Vertebrates of the wet tropics rainforests of Australia: species distributions and biodiversity.’ (Cooperative Research Centre for Tropical Rainforest Ecology and Management: Cairns.)
Winter, J. W., Dillewaard, H. A., Williams, S. E., and Bolitho, E. E. (2004). Possums and gliders of north Queensland: distribution and conservation status. In ‘The Biology of Australian Possums and Gliders’. (Eds. R. L. Goldingay, S. M. Jackson.) pp. 26–50. (Surrey Beatty and Sons: Sydney.)
Yang, Z. (1996). Among-site rate variation and its impact on phylogenetic analyses. Trends in Ecology & Evolution 11, 367–372.
| Among-site rate variation and its impact on phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar |