VISA and hVISA in hospitals
Iain B GosbellA Ingham Institute for Applied Medical Research
Liverpool, NSW, Australia
B School of Medicine
University of Western Sydney
Penrith, NSW, Australia
C Department of Microbiology and Infectious Diseases
Sydney South West Pathology Service
Liverpool, NSW, Australia
Tel: +61 2 8738 9024
Fax: +61 2 9602 9441
Email: i.gosbell@uws.edu.au
Microbiology Australia 35(1) 29-34 https://doi.org/10.1071/MA14009
Published: 17 February 2014
Staphylococcus aureus is noted for its clinical spectrum of disease ranging from asymptomatic colonisation to overwhelming sepsis and death and for its ability to become resistant to antibiotics. Resistance to beta-lactams, methicillin resistance, was first described 50 years ago, becoming a clinical problem in hospitals in the 1970s and the community in the 1990s. MRSA strains that originated in hospitals are usually also resistant to most of the non-beta-lactams as well, leaving vancomycin as the main parenteral drug to treat serious MRSA infections, with the role of new drugs like daptomycin and linezolid not well defined. MRSA strains can exhibit low-level resistance to vancomycin (vancomycin-intermediate S. aureus [VISA]), probably due to a thickened cell wall, which results in the trapping of vancomycin away from the active site of the septum in dividing cells. Detecting this resistance is difficult as multiple genetic pathways lead to this resistance, obviating a molecular test, forcing reliance on phenotypic tests, all of which have issues with sensitivity, specificity and cost. Mortality of bloodstream infection correlates with vancomycin MIC so in this situation the MIC should be determined by Etest or microbroth dilution especially if endocarditis is present. Detection of resistant subpopulations (heterogeneous vancomycin-intermediate S. aureus [hVISA]) can be done with the expensive and time-consuming population analysis profile (PAP) but it is unclear if this confers additional therapeutic information.
Types of vancomycin resistance in Staphylococcus aureus
There are two essential types of reduced vancomycin susceptibility in S. aureus: (1) that conferred by cell wall thickening resulting in a small rise in the MIC and/or the presence of subpopulations with a modestly elevated MIC1; and (2) that conferred by horizontal gene transfer of the vanA gene complex from vancomycin-resistant enterococci (VRE) to S. aureus, causing a more marked rise in the vancomycin MIC and clinical failure2.
The Clinical and Laboratory Standards Institute (CLSI) reduced the breakpoints for vancomycin resistance in 2006 in response to the evidence that modest elevations of MIC were associated with a reduced likelihood of clinical response, and currently define vancomycin-susceptible S. aureus (VSSA) strains as having a MIC ≤4 mg/L, VISA as having MICs 4–8mg/L, and vancomycin-resistant S. aureus (VRSA) strains as having an MIC ≥161. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) defines VSSA as strains with an MIC ≤2 and VRSA as having an MIC ≥4, with no intermediate division1.
The first described clinical isolate of VISA was the so-called ‘Mu50’ strain, isolated from a child with an MRSA sternotomy infection that failed to respond to vancomycin but responded to debridement and arbekacin plus ampicillin/sulbactam; the microbroth vancomycin MIC was 8 mg/L3.
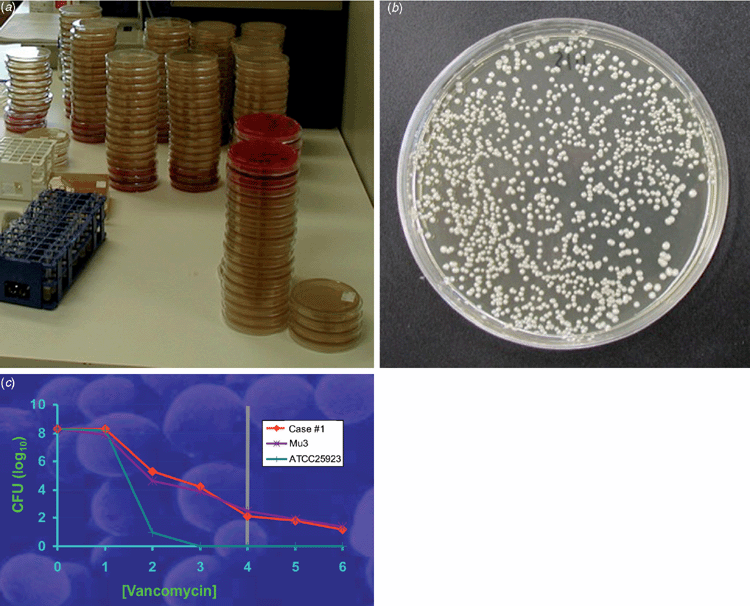
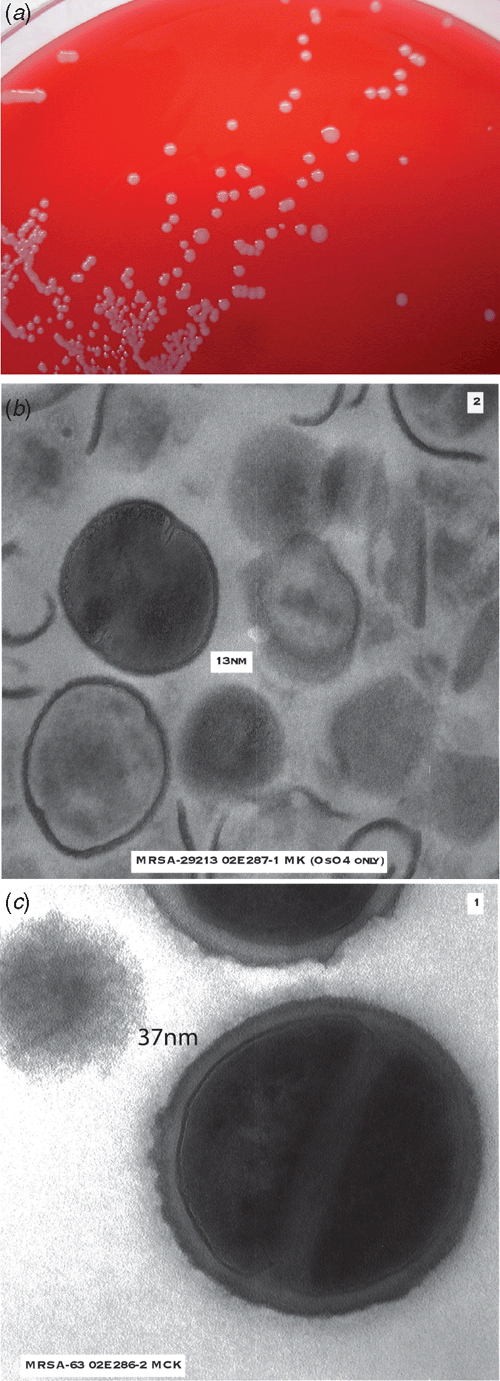
Mu50 and subsequent VISA isolates exhibit common characteristics (Figure 1)1. They grow more slowly than VSSA, exhibit pleomorphic colonial morphology, have reduced or delayed coagulase, demonstrate thickened cell walls on electron microscopy, have alterations in cell wall metabolism, reduced susceptibility to lysostaphin and decreased autolysis.
|
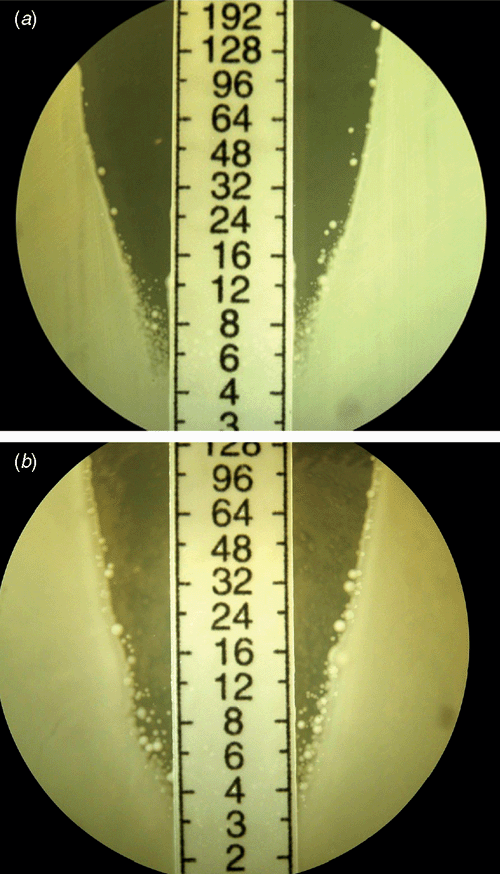
hVISA strains usually have a vancomycin MIC in the CLSI and EUCAST susceptible ranges but possess subpopulations with a raised MIC, detectable by performing a PAP, where serial 10-fold dilutions of a suspension of S. aureus are plated on a series of agar plates containing increasing dilutions of vancomycin1. This was first described by Hiramatsu’s group on the so-called ‘Mu3’ strain4, and Wootton et al. refined this by using a series of plates with doubling concentrations of vancomycin and serial 10-fold dilutions of the inocula with Mu3 as the control, generating the categories of VSSA (PAP-AUC ratio test: Mu3 <0.9), hVISA (ratio 0.9–1.35) and VISA (ratio >1.35) (Figure 2)5.
Just over a dozen patients to date have had MRSA isolated that possess the vanA gene complex2. Most are US origin; none have been reported from Australasia to date. The US patients are older with multiple co-morbidities, were colonised with vanA VRE and had high inoculum infections at sites difficult to penetrate with systemically administered vancomycin to which they have been extensively exposed. The vanA genes appear to have moved via plasmids from VRE into MRSA, conferring high-level resistance to vancomycin, therapeutic failure and major infection control issues.
Mechanism of resistance in VISA/hVISA
Hiramatsu has proposed the affinity-trapping hypothesis to explain vancomycin resistance in VISA/hVISA strains6. VISA/hVISA strains increase the cell wall thickness and vancomycin this binds to the outer layers of the cell wall and it is not available to penetrate into the deeper layers and thus interfere with cell division.
VISA/hVISA strains do not possess any elements of the vanA, vanB,vanC, etc., gene complexes found in enterococci that confer vancomycin resistance1. Many isolates have mutations in the walKR operon, which results in low activity, reduced autolysis and an increase in cell wall thickening1. The mprF/fmtC mutations result in reduced cell wall cross-linking1. There are also mutations in the graR genes and possible vraR as well, which results in upregulation of the cell wall stimulon1.
Mwangi et al. applied whole genome sequencing to five sequential MRSA human blood culture isolates and found 35 point mutations across the five isolates, including genes involved in the cell envelope stress response, mutations in the agr quorum sensing system, the walKR wall operon and genes involved in peptidoglycan biosynthesis7. The group also detected mprFyycG, rpoB and rpoC gene mutations that were associated with daptomycin resistance (the patient was naïve to this drug). Cameron et al., performed comparative genomics in a seven sets of vancomycin exposed isogenic S. aureus pairs and found that serine/threonine phosphatase stp1 contributes to reduce vancomycin susceptibility to vancomycin, in addition to previously described mutations such as vraG, agaR, yvqF and rpoB8.
Clinical significance of VISA and hVISA
A recent systematic review and meta-analysis of the clinical significance of the vancomycin MIC in MRSA bloodstream infections compared mortality where the MIC was ≥1.5 µg/mL to isolates where the MIC was <1.5 µg/mL and found an odds ratio of death of 1.64 (95% CI 1.14–2.37) in the higher MIC group9.
Holmes et al. compared patients with MRSA bloodstream infection where the vancomycin MIC was 2.0 or 3.0 versus MIC of ≤1.5 and found an all-cause 30-day mortality of 27% versus 12.5% (P < 0.001). The association with vancomycin MIC also occurred within the groups treated with flucloxacillin and in those with methicillin-susceptible S. aureus; the authors suggested that the raised vancomycin MIC might be an epiphenomenon10.
Currently the clinical significance of hVISA status is unclear. van Hal and Paterson performed a systematic review and meta-analysis of the clinical significance of hVISA11. They found hVISA infections had a clinical failure rate 2.37 times that of those with VSSA (95% CI 1.53–3.67), but all-cause 30-day mortality in the two groups was not significantly different. Peleg et al. described the greater wax moth (Galleria mellonella) model in which S. aureus strains with reduced susceptibility to vancomycin were associated with decreased pathogenicity and this correlated with decreased agr functional status12.
Testing for VISA and hVISA
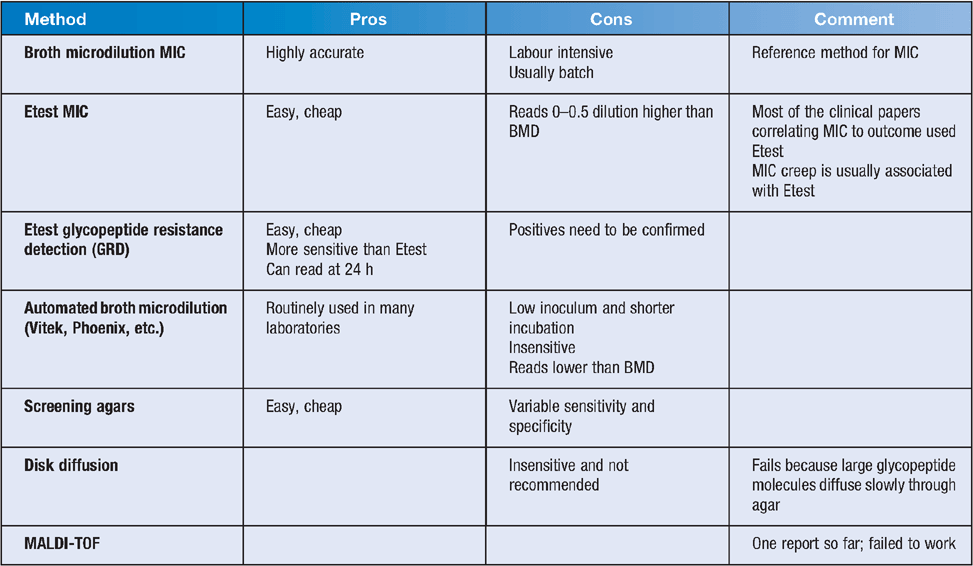
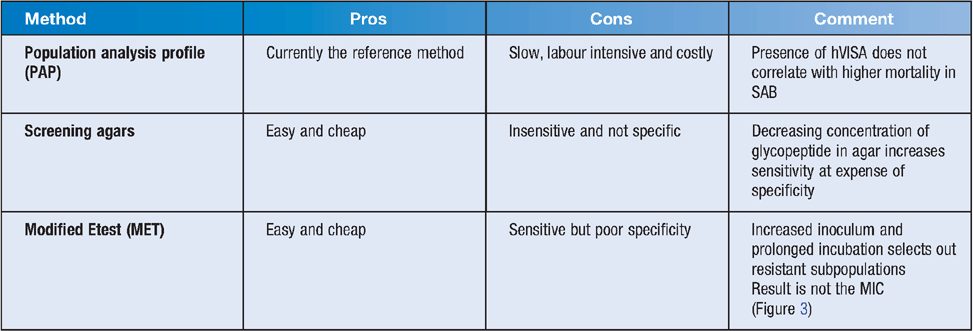
Testing can be done using a number of methods, as summarised in Tables 1 and 2.
|
|
|
Howden et al. in a comprehensive review outlined possible approaches to detection of VISA/hVISA1. If laboratory testing for VISA/hVISA is readily available, they suggest a laboratory-based approach, in which a vancomycin broth MIC could be used or alternatively modified Etest or an Etest GRD with confirmation of positives with a PAP, and if reduced susceptibility to vancomycin was demonstrated the treating doctor should consider daptomycin or linezolid. If VISA/hVISA testing is not readily available, a clinical approach could be adopted where a tardy clinical response might prompt the use of alternative agents such as daptomycin or linezolid. In clinical practice in the treatment of MRSA bacteraemia clinical failure often prompts the use of these agents before results of VISA/hVISA testing are available.
van Hal et al. compared different testing methodologies for detection of hVISA14. Four hundred and fifty-eight consecutive MRSA blood culture isolates were tested with PAP, macromethod Etest, glycopeptide resistance detection, GRD Etest and vancomycin MICs by Etest, by broth microdilution using CLSI criteria and Vitek2. Four hundred and fifty-eight isolates from 470 episodes of blood stream infection were analysed; 55 were hVISA and four were VISA by PAP, the latter 4 were excluded. The sensitivity and specificity of the various methods compared with PAP were Etest 91% and 66%, broth microdilution 89% and 84%, modified Etest 89 and 55%, broth microdilution (cutoff ≥2 mg/L) 82% and 97%, Etest (cutoff ≥2 mg/L) 71% and 94%, GRD Etest 71% and 94%, and the Vitek2 (MIC estimate ≥2 mg/L) 25% and 96%, respectively. PAP, modified Etest, GRDE test, broth microdilution, standard Etest and Vitek2 cost $70–$320, $29, $12, $8–$38, $15 and $18 per isolate, respectively. Etest tended to read higher and Vitek2 lower than BMD. Performing PAP on all the isolates would have 100% sensitivity and specificity (assuming PAP is the gold standard) but cost $32,000 to test all MRSA isolates in the study. Screening with Etest with cutoff MIC ≥1.5 confirming positives with PAP gave 98.9% sensitivity and cost ~$20,000. MET followed by PAP gave a sensitivity of 89% but cost ~$30,000. Other test combinations were cheaper but insensitive.
Holmes et al. recently summarised key points about the clinical and laboratory implications of the different types of vancomycin-reduced susceptibility15. For methicillin-susceptible S. aureus infections, β-lactams give better results than glycopeptides. For invasive MRSA infections, optimal vancomycin dosing and source control are pivotal. In this situation an MIC should be determined, preferably by BMD but Etest was satisfactory. If there was clinical failure of vancomycin and/or one of the recommended methods suggested reduced vancomycin susceptibility, daptomycin or linezolid should be considered. If the MIC ≥16 mg/L polymerase chain reaction testing for vanA and vanB genes should be performed as the isolate may be a VRSA.
MIC creep
van Hal and Fowler summarised the studies looking at MIC creep, which refers to the slow increase in the MIC of the MRSA isolates over period of time13. Creep is mostly demonstrated if Etest is used and not found in most studies using broth microdilution. Studies that demonstrated MIC creep tended to be single-centre studies. Typing was only performed in a small number of studies and where this was done, creep was demonstrated where there was a clonal emergence of strains with increased MICs, which probably explains the phenomenon.
Are VISA/hVISA strains less susceptible to daptomycin?
Daptomycin is a large molecular weight compound with a site of action in the cell wall, like vancomycin, and it has been hypothesised that VISA/hVISA strains might be less susceptible to daptomycin. A patient treated unsuccessfully with vancomycin but never treated with daptomycin, has a series of MRSA isolates from blood demonstrating increasing resistance to vancomycin and also daptomycin16. Sakoulas et al. found MRSA strains that were heterogeneously resistant to vancomycin were also heterogeneously resistant to daptomycin17. Another group found MRSA MICs to daptomycin and vancomycin were strongly correlated (P > 0001, χ2 test)18.
Conclusions
If there is clinical failure of glycopeptide treatment of MRSA infection, especially bacteraemia, generally daptomycin or linezolid will be selected before screening or definitive testing for VISA/hVISA can be performed. In the case of MRSA bacteraemia, especially if endocarditis or other deep focus is present, the MIC should be determined, either by Etest (not modified Etest which is a screen for hVISA) or broth microdilution.
Fortunately most MRSA infections do not involve bacteraemia. Vancomycin is cheap, generally well tolerated, has defined therapeutic drug monitoring strategies, and works well for the parenteral treatment of most MRSA infections.
Future directions
The clinical significance of hVISA needs to be defined. A cheap reliable test that predicts clinical failure with vancomycin is needed. The role of daptomycin, linezolid and the new anti-MRSA cephalosporin ceftaroline in serious MRSA infections needs to be clarified.
Acknowledgements
The author acknowledges the NSW Ministry of Health and the former Health Research Foundation, Sydney South West for funding the early work on hVISA/VISA research, staff of Sydney South West Pathology Service (Liverpool) who performed much of the laboratory work (especially Thelma Barbagiannakos, Yvonne Kwok, Jo Mercer, Dehua Chen, and Michael Wehrhahn), Murray Killingsworth of the Anatomical Pathology Department of SSWPS who provided various electron micrographs, Peter Ward from the Austin Hospital in Melbourne who trained Thelma in how to perform PAP and for helpful advice on the laboratory detection of hVISA/VISA, and members of the Antibiotic Resistance and Mobile Elements Group (ARMEG) (Slade Jensen, Bjorn Espedido and Sebastiaan van Hal) who are continuing the research on resistance in MRSA.
References
[1] Howden, B.P. et al. (2010) Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23, 99–139.| Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXks1Sktb8%3D&md5=a6a17476112da9d497f3708c10668e7fCAS | 20065327PubMed |
[2] Perichon, B. and Courvalin, P. (2009) VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53, 4580–4587.
| VanA-type vancomycin-resistant Staphylococcus aureus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtl2itLvF&md5=a133feef78b60620864ae466a5247101CAS | 19506057PubMed |
[3] Hiramatsu, K. et al. (1997) Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40, 135–136.
| Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXltVCit7w%3D&md5=63fcf86212e15251f1ffb60f911f4eedCAS | 9249217PubMed |
[4] Hiramatsu, K. et al. (1997) Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350, 1670–1673.
| Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXns1em&md5=afcc9b21aa311ab1390f7cba53540786CAS | 9400512PubMed |
[5] Wootton, M. et al. (2001) A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47, 399–403.
| A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXjtVamtLk%3D&md5=9bca4e07bd879a074dfc727afed44116CAS | 11266410PubMed |
[6] Hiramatsu, K. (2001) Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1, 147–155.
| Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XltVOnsbs%3D&md5=378b6014be993f76c393af03b38d4559CAS | 11871491PubMed |
[7] Mwangi, M.M. et al. (2007) Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 104, 9451–9456.
| Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmtlegu7g%3D&md5=de43bb7acfd1cf49161977bd54fa0d10CAS | 17517606PubMed |
[8] Cameron, D.R. et al. (2012) Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J. Infect. Dis. 205, 1677–1687.
| Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xnt1amtLk%3D&md5=a9c35de4cb01beed690216d605c959aaCAS | 22492855PubMed |
[9] van Hal, S.J. et al. (2012) The clinical significance of vancomycin minimum inhibitory concentration in Staphylococcus aureus infections: a systematic review and meta-analysis. Clin. Infect. Dis. 54, 755–771.
| The clinical significance of vancomycin minimum inhibitory concentration in Staphylococcus aureus infections: a systematic review and meta-analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjtFKjtLg%3D&md5=ced5af9e0145acb453fb591f08b18c69CAS | 22302374PubMed |
[10] Holmes, N.E. et al. (2011) Antibiotic choice may not explain poorer outcomes in patients with Staphylococcus aureus bacteremia and high vancomycin minimum inhibitory concentrations. J. Infect. Dis. 204, 340–347.
| Antibiotic choice may not explain poorer outcomes in patients with Staphylococcus aureus bacteremia and high vancomycin minimum inhibitory concentrations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXovVKqt7Y%3D&md5=173e41384998a38445f6a292e76f35bdCAS | 21742831PubMed |
[11] van Hal, S.J. and Paterson, D.L. (2011) Systematic review and meta-analysis of the significance of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 55, 405–410.
| Systematic review and meta-analysis of the significance of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXisVegsLs%3D&md5=50acbb1098da587e2ea787e40512a049CAS | 21078939PubMed |
[12] Peleg, A.Y. et al. (2009) Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J. Infect. Dis. 199, 532–536.
| Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection.Crossref | GoogleScholarGoogle Scholar | 19125671PubMed |
[13] van Hal, S.J. and Fowler, V.G. (2013) Is it time to replace vancomycin in the treatment of Methicillin-Resistant Staphylococcus aureus infections? Clin. Infect. Dis. 56, 1779–1788.
| Is it time to replace vancomycin in the treatment of Methicillin-Resistant Staphylococcus aureus infections?Crossref | GoogleScholarGoogle Scholar | 23511300PubMed |
[14] van Hal, S.J. et al. (2011) Performance of various testing methodologies for detection of heteroresistant vancomycin-intermediate Staphylococcus aureus in bloodstream isolates. J. Clin. Microbiol. 49, 1489–1494.
| Performance of various testing methodologies for detection of heteroresistant vancomycin-intermediate Staphylococcus aureus in bloodstream isolates.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtFKrt7fJ&md5=56d9e77e39da14d3220b9148c0761ceeCAS | 21270232PubMed |
[15] Holmes, N.E. et al. (2012) Relationship between vancomycin-resistant Staphylococcus aureus, vancomycin-intermediate S. aureus, high vancomycin MIC, and outcome in serious S. aureus infections. J. Clin. Microbiol. 50, 2548–2552.
| Relationship between vancomycin-resistant Staphylococcus aureus, vancomycin-intermediate S. aureus, high vancomycin MIC, and outcome in serious S. aureus infections.Crossref | GoogleScholarGoogle Scholar | 22593595PubMed |
[16] van Hal, S.J. et al. (2011) Emergence of daptomycin resistance following vancomycin-unresponsive Staphylococcus aureus bacteraemia in a daptomycin-naive patient--a review of the literature. Eur. J. Clin. Microbiol. Infect. Dis. 30, 603–610.
| Emergence of daptomycin resistance following vancomycin-unresponsive Staphylococcus aureus bacteraemia in a daptomycin-naive patient--a review of the literature.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3MvivVyhug%3D%3D&md5=709e77c977c5ef577242570ca66c4d81CAS | 21191627PubMed |
[17] Sakoulas, G. et al. (2006) Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 50, 1581–1585.
| Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjvFOrtrc%3D&md5=55dfb7faf99b61c7e5a329ea5f84a54dCAS | 16569891PubMed |
[18] Patel, J.B. et al. (2006) An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin. Infect. Dis. 42, 1652–1653.
| An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XlvVOrsrg%3D&md5=65e9bddf806607fa511204d6b26d0bdcCAS | 16652325PubMed |
Biography
Professor Iain Gosbell is the Foundation Professor of Infectious Diseases and Microbiology at the School of Medicine, University of Western Sydney, Australia. He described the emergence of community MRSA in south western Sydney, and subsequently developed research interests in the epidemiology of MRSA in the community and also in hospitals, and the detection of vancomycin resistance in MRSA. He established the Infection Prevention Unit at Liverpool Hospital in conjunction with their Infection Control Practitioners. Since commencing at UWS, he helped establish the Antibiotic Resistance and Mobile Elements Group which is now based at the Ingham Institute for Applied Medical Research at Liverpool. ARMEG consists of Professor Gosbell, Dr Slade Jensen, Dr Björn Espedido and Associate Professor Sebastiaan van Hal. Its mission is to research the genetics responsible for antibiotic resistance, especially vancomycin and daptomycin resistance in MRSA, plasmid biology of MRSA, and the contribution of bacterial biofilms to healthcare-associated infections. Professor Gosbell also promotes the importance of Microbiology and Infectious Diseases and the use of Information Technology in Medical Education. He was awarded the 2013 ASM bioMérieux Identifying Resistance Award.