Influenza vaccine production technologies: past, present and future
Yingxia Wen A and Ethan C Settembre A BA Seqirus, A CSL Company
50 Hampshire Street
Cambridge, MA 02139, USA
B Tel: 1 617 254 5354 Email: Ethan.Settembre@Seqirus.com
Microbiology Australia 38(2) 52-54 https://doi.org/10.1071/MA17026
Published: 21 March 2017
Influenza is a constantly evolving global health threat that leads to substantial morbidity and mortality particularly in vulnerable populations at either end of the age spectrum. Society has responded by creating a global public-private system that involves constant surveillance, candidate virus generation, and release reagent generation linked to worldwide influenza vaccine manufacturing capabilities. It was initially recognised that influenza circulates as multiple antigenically distinct subtypes, which led to the generation of vaccines containing multiple influenza strains. The first and still current major process used for influenza vaccine production is infection of embryonated hen’s eggs with influenza virus. While this approach was a true advancement, some shortcomings such as lack of vaccine match to circulating strains due to egg adaptation and production capacity limitations have led to recent innovations in mammalian cell production and synthetic technologies aimed at further improving global influenza responses.
Influenza viruses are a constant threat to human health with seasonal epidemics responsible for 250 000 to 500 000 deaths worldwide and over three million cases globally each year. Fear that a pandemic such as the 1918 pandemic where ~50 million individuals died, has led to continued efforts to improve influenza vaccine production1. Influenza poses such a threat due to a high mutation rate resulting in seasonal changes in virus antigenicity (antigenic drift), the presence of multiple co-circulating antigenically distinct subtypes, animal reservoirs where antigenically distant viruses can reside that may contain new gene segment combinations (antigenic shift), and high aerosol transmissibility.
The Global Influenza Surveillance and Response System (GISRS) has been designed to address these biological characteristics2. This system currently involves: 143 WHO designated National Influenza Centers (NICs) where original clinical specimens are collected and partially analysed; 6 WHO Collaborating Centers (WHO CCs) that antigenically characterise collected viruses and generate candidate vaccine viruses for vaccine production; and 4 Essential Regulatory Laboratories (ERLs) that generate and distribute vaccine release reagents. This complicated international system has commercial manufacturers as close partners who use the WHO recommended viruses to generate safe and efficacious influenza vaccines. The goal of the manufacturing processes is to obtain large quantities of properly folded hemagglutinin (HA), the primary vaccine antigen.
Inactivated influenza vaccines (IIVs) were first generated in the 1940s using embryonated hen’s eggs (Figure 1). The upstream manufacturing process involves virus propagation in eggs requiring virus inoculation, egg incubation, egg candling and allantoic fluid harvest from millions of high quality fertilised eggs. This process represented an advance at the time and still serves as the major manufacturing platform worldwide. However, the upstream process can be inefficient and labor intensive with steps that are open and thus susceptible to microbial contamination which can occasionally lead to vaccine batch rejection.
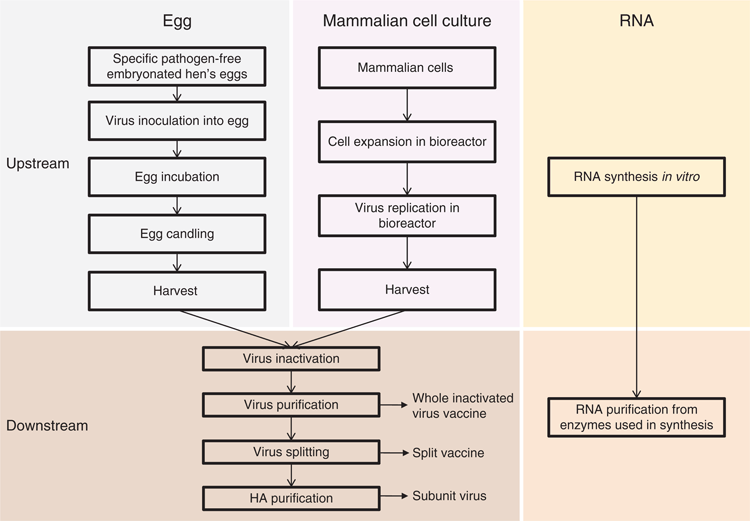
|
Egg passage of virus has been shown to alter the HA genetic sequence that can at times change the HA antigenicity from that of the wild-type virus3,4. The normal role of HA is to bind a cellular receptor and mediate viral-cell fusion. HA genetic mutations are largely due to selective pressures imposed when a virus that has adapted to use receptors found on human respiratory epithelium cells (α2,6-linked sialosides) needs to readapt to those found on egg allantoic cavity cells (α2,3-linked sialosides). Immune responses to the altered HA may not inhibit infection of the circulating virus to which the individual may be exposed, thus potentially reducing vaccine effectiveness5.
Mammalian cell culture has been shown to function effectively for IIV production both simplifying early steps and allowing for quicker vaccine production initiation because mammalian cells can be rapidly expanded in bioreactors (Figure 1). Generation of a mammalian cell-based vaccine in an aseptic and controlled environment also reduces possibility for contamination and removes the reliance upon eggs. Viruses that have been entirely propagated in mammalian cells have been reported to be more representative of wild-type viruses6,7. Overall, mammalian cell-based influenza vaccines provide equivalent or better protection than egg-based vaccine in animal models8,9 and are shown to be both safe and effective in clinical trials10.
The IIV manufacturing process still has several limitations: production requires a prolonged process to generate vaccine seed viruses, virus needs to be produced in large quantity, and the downstream process is complex (Figure 1). Immunologic reagents are also needed for antigen quantification and vaccine release11. Live-attenuated vaccines and recombinant vaccines have shown some potential advantages over inactivated vaccines. For example, live attenuated vaccines can elicit additional cellular immune response and less virus is needed for vaccine production. However, current live-attenuated vaccines are still generated from hen’s egg, which may result in antigenic changes during production12. Recombinant vaccines can be designed to match the circulating virus much like mammalian cell production processes when a cell-based seed virus is used, however, purification of the metastable HA can be challenging and currently higher doses appear to be required to achieve similar immunogenicity as IIVs.
The limitations of the current IIVs and the current processes provide a strong impetus for next-generation vaccine technology development. One such process improvement has been to use synthetic biology to generate a synthetic vaccine seed virus. It has been demonstrated this process is quick with a synthetic vaccine seed virus being generated within a week of a target virus identification13. The technique uses enzymatic, cell-free gene assembly to synthesise hemagglutinin and neuraminidase genes from sequence, and then the vaccine seed virus is produced when mammalian cells are transfected with both antigen expression constructs and plasmid DNAs encoding viral backbone genes. Improved vaccine virus backbones can further increase vaccine yield and process robustness.
RNA vaccine technologies promise another potential system advancement. Synthetic mRNA influenza vaccines can be produced quickly on small manufacturing footprints with high yields, leading to lower cost of goods (COGs). The SAM® influenza vaccine technology is based on self-amplifying mRNA delivered by a synthetic lipid nanoparticle (LNP)14. The vaccine production process includes rapid and accurate enzymatic, cell-free mRNA synthesis followed with a simple purification and formulation process. During the initial H7N9 influenza outbreak, SAM influenza vaccine technology was used to generate a vaccine candidate in 8 days. The vaccine was shown to elicit potent immune responses in mice, comparable with inactivated vaccine15. If this vaccine platform proves safe and potent in humans, fully synthetic vaccine technologies could provide unparalleled speed of response and potentially greater vaccine effectiveness. From current to future technologies, improvements in influenza vaccine production are essential to enable improved responses in seasonal epidemics and pandemics.
References
[1] Poland, G.A. et al. (2001) Influenza vaccines: a review and rationale for use in developed and underdeveloped countries. Vaccine 19, 2216–2220.| Influenza vaccines: a review and rationale for use in developed and underdeveloped countries.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3MvgvFSmtA%3D%3D&md5=0446f485cc4242f18e6949fec8f62a9cCAS |
[2] Pereyaslov, D. et al. (2016) Improving the representativeness of influenza viruses shared within the WHO Global Influenza Surveillance and Response System. Influenza Other Resp. Viruses 10, 68–75.
| Improving the representativeness of influenza viruses shared within the WHO Global Influenza Surveillance and Response System.Crossref | GoogleScholarGoogle Scholar |
[3] Oxford, J.S. et al. (1991) Direct isolation in eggs of influenza A (H1N1) and B viruses with haemagglutinins of different antigenic and amino acid composition. J. Gen. Virol. 72, 185–189.
| Direct isolation in eggs of influenza A (H1N1) and B viruses with haemagglutinins of different antigenic and amino acid composition.Crossref | GoogleScholarGoogle Scholar |
[4] Robertson, J.S. et al. (1985) Alterations in the hemagglutinin associated with adaptation of influenza B virus to growth in eggs. Virology 143, 166–174.
| Alterations in the hemagglutinin associated with adaptation of influenza B virus to growth in eggs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2MXitVSqt7c%3D&md5=4534dab6adba72c97623668f8aa8acbfCAS |
[5] Wood, J.M. et al. (1989) H1N1) vaccine efficacy in animal models is influenced by two amino acid substitutions in the hemagglutinin molecule. Virology 171, 214–221.
| H1N1) vaccine efficacy in animal models is influenced by two amino acid substitutions in the hemagglutinin molecule.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1MXkvFGjtbo%3D&md5=e44e0b4b653b0135273aff98ffe89f9cCAS |
[6] Rocha, E.P. et al. (1993) Comparison of 10 influenza A (H1N1 and H3N2) haemagglutinin sequences obtained directly from clinical specimens to those of MDCK cell- and egg-grown viruses. J. Gen. Virol. 74, 2513–2518.
| Comparison of 10 influenza A (H1N1 and H3N2) haemagglutinin sequences obtained directly from clinical specimens to those of MDCK cell- and egg-grown viruses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2cXisVOgtg%3D%3D&md5=0087e02d661c6ad1a11e032a6ccde2cdCAS |
[7] Wang, M.L. et al. (1989) Extensive heterogeneity in the hemagglutinin of egg-grown influenza viruses from different patients. Virology 171, 275–279.
| Extensive heterogeneity in the hemagglutinin of egg-grown influenza viruses from different patients.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1MXkslKjtLc%3D&md5=ce164f71d25dd791522f15ad305d8b2dCAS |
[8] Katz, J.M. and Webster, R.G. (1989) Efficacy of inactivated influenza A virus (H3N2) vaccines grown in mammalian cells or embryonated eggs. J. Infect. Dis. 160, 191–198.
| Efficacy of inactivated influenza A virus (H3N2) vaccines grown in mammalian cells or embryonated eggs.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL1MzktFCmtA%3D%3D&md5=04440e945896597f05fa9de28b3102b5CAS |
[9] Alymova, I.V. et al. (1998) Immunogenicity and protective efficacy in mice of influenza B virus vaccines grown in mammalian cells or embryonated chicken eggs. J. Virol. 72, 4472–4477.
| 1:CAS:528:DyaK1cXis1Kmtr0%3D&md5=b993d044a992ffd06c18077b0cbfa614CAS |
[10] Frey, S. et al. (2010) Clinical efficacy of cell culture-derived and egg-derived inactivated subunit influenza vaccines in healthy adults. Clin. Infect. Dis. 51, 997–1004.
| Clinical efficacy of cell culture-derived and egg-derived inactivated subunit influenza vaccines in healthy adults.Crossref | GoogleScholarGoogle Scholar |
[11] Settembre, E.C. et al. (2014) Bringing influenza vaccines into the 21st century. Hum. Vaccin. Immunother. 10, 600–604.
| Bringing influenza vaccines into the 21st century.Crossref | GoogleScholarGoogle Scholar |
[12] Jin, H. and Chen, Z. (2014) Production of live attenuated influenza vaccines against seasonal and potential pandemic influenza viruses. Curr. Opin. Virol. 6, 34–39.
| Production of live attenuated influenza vaccines against seasonal and potential pandemic influenza viruses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtVarsLnN&md5=a8401958eaac7dbe772e869904be008bCAS |
[13] Dormitzer, P.R. et al. (2013) Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci. Transl. Med. 5, 185ra68.
| Synthetic generation of influenza vaccine viruses for rapid response to pandemics.Crossref | GoogleScholarGoogle Scholar |
[14] Geall, A.J. et al. (2012) Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 109, 14604–14609.
| Nonviral delivery of self-amplifying RNA vaccines.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVaqu7zI&md5=d33f67e640e92a8fd96087247767a391CAS |
[15] Hekele, A. et al. (2013) Rapidly produced SAM((R)) vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2, e52.
| Rapidly produced SAM((R)) vaccine against H7N9 influenza is immunogenic in mice.Crossref | GoogleScholarGoogle Scholar |
Biographies
Dr Yingxia Wen is Head of Biochemistry in Seqirus, with eight years of research and development in viral vaccines. She has led the development of influenza, HIV, RSV and CMV vaccines during her time at Novartis Vaccines and Seqirus. Before joining Novartis Vaccines, Yingxia worked in Protein Biochemistry group of Wyeth Research with increasing responsibility. Yingxia received her PhD in Biochemistry from Rutgers University and postdoctoral training from Dr Tom Maniatis Laboratory at Harvard University.
Dr Ethan Settembre is the Vice President, Head of Research for Seqirus. He holds a PhD in biochemistry from Cornell University and completed his post-doctoral training in Structural Virology at Harvard Medical School. He then joined Novartis Vaccines and Diagnostics in 2008 where he held several key positions in Research developing vaccines against multiple viral targets including influenza. Currently he heads the Research team in Seqirus focused on influenza vaccine development.


