Diagnosis of dermatophytes: from microscopy to direct PCR
Sarah E. Kidd A and Gerhard F. Weldhagen AA National Mycology Reference Centre, Microbiology and Infectious Diseases, SA Pathology, Frome Road, Adelaide, SA 5000, Australia.

Sarah Kidd is the Head of the National Mycology Reference Centre at SA Pathology, as well as Adjunct Senior Lecturer at the University of Adelaide. She completed a Bachelor of Medical Science with Honours (1999) and a PhD in Medicine (2003), both at the University of Sydney, and has undertaken postdoctoral training and research at the University of British Columbia, Vancouver, Canada, and at Monash University–The Alfred Hospital, Melbourne. She is a Fellow of the Australian Society for Microbiology (FASM), Secretary of the Australia and New Zealand Mycoses Interest Group (ANZMIG), convenes the biennial Mycology Masterclass, and provides mycology expertise to the Royal College of Pathologists of Australasia Quality Assurance Programs. |

Gerhard Weldhagen is the supervising pathologist for the National Mycology Reference Centre at SA Pathology. He completed his undergraduate studies in 1993, receiving a Bachelor’s degree in Medicine and Surgery (MBChB) from the University of Pretoria, South Africa. This was followed by a Master’s degree in Clinical Microbiology (MMed (Path) cum laude) and subsequently a PhD in Microbiology, conferred by the University of Pretoria in 2002 and 2005 respectively. After settling in Australia during 2009, a Fellowship of the Royal College of Pathologists of Australasia (FRCPA) was attained during the same year. Current interests include the role of molecular assays in diagnostic microbiology, including mycology. |
Microbiology Australia 43(1) 9-13 https://doi.org/10.1071/MA22005
Submitted: 31 December 2021 Accepted: 5 February 2022 Published: 8 April 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Dermatophyte fungi are a common cause of skin, nail and hair infections globally, ranging from mild to cosmetically disfiguring, or even invasive infections in rare cases. Specimens requiring fungal microscopy and culture for suspected dermatophyte infection make up a significant portion of the workload in diagnostic microbiology laboratories. Whilst still considered the gold standard, a dermatophyte culture-based method is labour intensive, has poor sensitivity, slow result turnaround time and requires significant expertise for identification of the fungi. Molecular diagnostics, especially real-time PCR, have the potential to improve diagnostic sensitivity, reduce labour requirements and decrease result turnaround times. Despite these advantages, a PCR-based approach may present some difficulties and disadvantages, most notably its diagnostic range and incompatibility with oral therapy prescribing requirements under the Pharmaceutical Benefits Scheme. Here we review current best practices and future prospects for laboratory diagnosis of dermatophyte infections, including the role of microscopy, culture and direct PCR.
Keywords: dermatophyte, Epidermophyton, fungal culture, medical mycology, microscopy, Microsporum, onychomycosis, real-time PCR, tinea pedis, Trichophyton.
Introduction
Dermatophyte fungi are a common cause of skin, nail and hair infections globally. The most common agents are Trichophyton rubrum and T. interdigitale,1–3 but other dermatophytes belonging to the genera Arthroderma, Trichophyton, Epidermophyton, Microsporum, Nannizzia, Paraphyton, and Lophophyton (the latter three genera formerly classified within Microsporum) also cause infection. Infections commonly present as tinea pedis and onychomycosis, but can affect any keratinised area of the body, having low clinical acuity. However, some infections may be debilitating or invasive in immunocompromised or elderly patients.4,5 The prevalence of onychomycosis is approximately 10% in the general population, but may increase to 50% in those aged >70 years.6 Infections are usually transmitted by direct or indirect human contact, but may also be acquired from animal sources or soil, depending on the species. The prevalence of these infections and the species that cause them appears to vary significantly by geographic region, and is well reviewed by Nowicka and Nawrot.3
Occasionally, yeasts such as Candida spp. and non-dermatophyte moulds such as Scopulariopsis spp., Aspergillus spp., Fusarium spp., and Acremonium spp. may cause onychomycosis, but the diagnosis is complicated by these fungi also being common environmental contaminants.7,8 Diagnostic guidelines require at least two subsequent isolations of these fungi in the absence of a dermatophyte and in the setting of direct microscopy exhibiting fungal hyphae not resembling dermatophytes.9
Effective antifungal agents are available for treatment of dermatophyte infections in topical or oral formulations, with the latter being necessary to treat refractory nail infections. However, for oral formulations to be prescribed through the Pharmaceutical Benefits Scheme (PBS), patients need to fulfil clinical requirements and laboratory diagnostic criteria including microscopy and dermatophyte culture.10 Therefore, these infections form a significant laboratory workload and reliable, sensitive, and specific diagnostic methods are needed for effective treatment.
While much of mycology continues to utilise conventional methods such as microscopy and culture, the diagnosis of dermatophyte infections is increasingly becoming modernised through the use of real-time PCR assays. Here we review current best practices and future prospects for laboratory diagnosis of dermatophyte infections, including the role of microscopy, culture and direct PCR, and Table 1 summarises the advantages and disadvantages of each.
|
Microscopy
Direct microscopy of specimens is an essential component of the diagnostic pathway for cutaneous fungal infections. It provides a relatively fast result demonstrating the presence or absence of fungal elements indicative of infection. More importantly, a skilled microscopist can differentiate dermatophyte hyphae from that of non-dermatophyte moulds and yeast pseudohyphae, and the budding yeast cells of Candida spp. from those of Malassezia spp. This alone may be sufficient for a diagnosis of dermatophyte and dermatophyte-like infections. However, observation of fungal elements does not necessarily indicate causation of infection or demonstrate viability of the fungal elements and it is not possible to determine the genus or species. Therefore, microscopy is a low-specificity diagnostic technique.
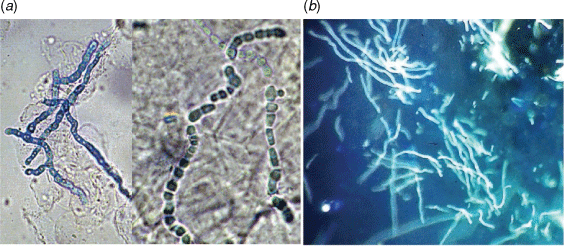
Traditionally, skin and nail specimens are digested in 10–20% potassium hydroxide (KOH) as a clearing agent for 3–16 h prior to microscopy, having a false-negative rate of 5–15%.11 Staining with chlorazol black E can provide additional contrast for visualising the fungal elements (Fig. 1a). However, optical brightener stains such as Calcofluor White or Blankophor, which bind to cellulose and chitin in the fungal cell wall and fluoresce under UV light (Fig. 1b), significantly improve the sensitivity of microscopy (82–91% vs 74–85% KOH preparation alone)12,13 and allows more rapid scanning of slides. Optical brightener stains are recommended in laboratory guidelines for dermatophyte studies.14
|
Culture
Culture remains the gold standard for diagnosis of dermatophyte and other fungal infections, yielding a specific aetiological agent that can be identified to species level. However, culture has low sensitivity,15 requires 2–4 weeks for growth, and potentially a further 2 weeks for identification. Species identification needs detailed examination of colony and microscopic morphology, requiring specialised media and significant expertise.
Culture media for dermatophyte studies should include a specialised isolation medium such as Lactritmel or Dermatophyte Test Medium, containing antibiotics and cycloheximide; this minimises growth of fast growing bacterial and fungal contaminants, allowing the slower growing dermatophyte to grow. Sabouraud’s dextrose agar containing antibiotics but not cycloheximide is also recommended for nail specimens to allow growth of potential causes of non-dermatophyte onychomycosis.
The identification of dermatophytes and other fungi requires specialised training and expertise, something of a dying art in the era of laboratory automation. Commercial matrix assisted laser desorption/ionisation-time of flight mass spectrometry (MALDI-ToF) databases have the potential to identify common dermatophyte species; however, many species, for example, Trichophyton benhamiae, T. equinum, T. erinacei, T. eriotrephon, T. interdigitale, T. mentagrophytes and T. tonsurans, cannot be reliably differentiated.16 Optimised protein extraction methodologies combined with supplemented databases may be far more accurate,17,18 although significant initial setup is required. DNA sequencing for species identification using the Internal Transcribed Spacer (ITS) rDNA region may be useful in some cases, but sequence databases contain numerous entries for dermatophytes with incorrect identifications that complicates making a reliable identification, and particularly between T. interdigitale and T. mentagrophytes.19
Real-time PCR assays
Given the slow and labour-intensive nature of conventional methods, many laboratories are looking towards real-time PCR for detection of dermatophytes in clinical samples.20–26 The primary advantage of this approach is the decreased turnaround time over culture, with results typically available within 1–3 days of receipt in the laboratory. Additionally, the sensitivity of PCR is around 20–30% higher than culture, in part because dermatophyte DNA can be detected even in the presence of fast-growing fungal contaminants that might overgrow the slower-growing dermatophytes in cultures.15,20,21,25 PCR-based identification reduces the need for morphologists, who could be utilised more effectively diagnosing fungi from life-threatening infections.
PCR assays may be commercial or in-house designed. In-house assays have the benefit of being customisable to existing laboratory platforms, as well as to include species that best represent the local epidemiology,23 and fungi involved in non-dermatophyte infections of the skin and nails. Commercial assays typically have the benefit of regulatory approval, lessening the validation requirements on the laboratory. The sensitivity of dermatophyte PCR assays has invariably been shown to exceed that of culture.20–26
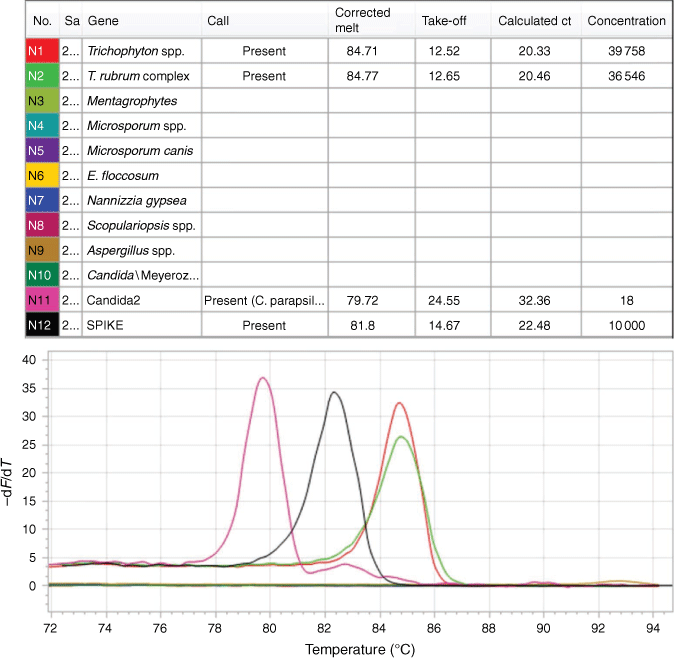
Some commercial assays include pan-dermatophyte detection and/or specific primers for detection of a limited number of species. For example, the Dermatophyte PCR Test (SSI Diagnostica, Denmark), utilises pan-dermatophyte primers targeting the CHS1 gene as well as T. rubrum-specific ITS2 primers, the DermaGenius Nail multiplex assay (PathoNostics, The Netherlands) contains species-specific primers for T. rubrum, T. interdigitale and C. albicans only, and the Dermatophytes Real-Time PCR (EurobioPlex, France) is designed to detect six dermatophyte species. The ‘Dermatophytes and other Fungi’ multiplex tandem PCR (MT-PCR) (AusDiagnostics, Sydney) detects at least 14 dermatophyte species, as well as four Candida species and two non-dermatophyte moulds, with species identification made possible by specific primers and melt curve analysis (Fig. 2). This assay does not currently differentiate all of the species that it detects,21,22 but differences in melt curve may be suggestive of certain species, requiring further validation. Given the limitations of species differentiation with PCR assays, it may only be possible to report the species complex, genus, or simply ‘dermatophyte detected’. This loss of species resolution limits insight into the potential origins of infection (i.e. anthropophilic vs zoophilic), but for most clinicians the faster turnaround time coupled with increased sensitivity, appears to be an acceptable trade-off.
PCR assays may not detect non-dermatophyte fungi that cause onychomycosis, which have an incidence of around 2–20%.7,8,27,28 The AusDiagnostics panel includes targets for Aspergillus spp., Scopulariopsis spp., and four Candida species but not Fusarium spp., Acremonium spp., and Neoscytalidium spp. Since these may be environmental contaminants or opportunistic skin flora, they may be detected in the absence of infection and should be reported with caution, potentially utilising reflex cultures where non-dermatophyte onychomycosis is suspected.
Real-time PCR detection of squalene epoxidase mutations conferring terbinafine resistance has recently been described.29 However, the significance and clinical utility of such assays are currently limited, as susceptibility breakpoints for dermatophytes remain tentative or unavailable.30
Despite the superior sensitivity of PCR compared to culture, dermatophyte detection in nail specimens requires confirmation by either microscopy or culture in order to satisfy Australian PBS requirements for prescription of oral terbinafine. This represents additional work, and does not always provide the necessary confirmation. Some of these cases may be attributed to the detection of DNA from non-viable dermatophytes (i.e. already inactivated by antifungal treatment).
Conclusions
PCR detection of dermatophyte infection brings many benefits to laboratories that are increasingly stretched for resources. However, currently, such advances come at the cost of accurate speciation, and the associated ecological and epidemiological information that accompanies it. Current PBS requirements and the increasingly apparent role of non-dermatophyte fungi in cutaneous infections means that there remains a role for microscopy and culture in routine diagnostic laboratories.
Data availability
Data sharing is not applicable as no new data were generated or analysed during this study.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This work did not receive any specific funding.
References
[1] Ghannoum, MA et al.. (2000) A large-scale North American study of fungal isolates from nails: the frequency of onychomycosis, fungal distribution, and antifungal susceptibility patterns. J Am Acad Dermatol 43, 641–648.| A large-scale North American study of fungal isolates from nails: the frequency of onychomycosis, fungal distribution, and antifungal susceptibility patterns.Crossref | GoogleScholarGoogle Scholar | 11004620PubMed |
[2] Ilkit, M and Durdu, M (2015) Tinea pedis: the etiology and global epidemiology of a common fungal infection. Crit Rev Microbiol 41, 374–388.
| Tinea pedis: the etiology and global epidemiology of a common fungal infection.Crossref | GoogleScholarGoogle Scholar | 24495093PubMed |
[3] Nowicka, D and Nawrot, U (2021) Tinea pedis-An embarrassing problem for health and beauty: a narrative review. Mycoses 64, 1140–1150.
| Tinea pedis-An embarrassing problem for health and beauty: a narrative review.Crossref | GoogleScholarGoogle Scholar | 34145648PubMed |
[4] Teo, TSP et al.. (2021) Mycetoma caused by Microsporum canis in a patient with renal transplant: a case report and review of the literature. Transpl Infect Dis 23, e13516.
| Mycetoma caused by Microsporum canis in a patient with renal transplant: a case report and review of the literature.Crossref | GoogleScholarGoogle Scholar | 33217133PubMed |
[5] Wang, R et al.. (2021) Invasive dermatophyte infection: a systematic review. Mycoses 64, 340–348.
| Invasive dermatophyte infection: a systematic review.Crossref | GoogleScholarGoogle Scholar | 33217082PubMed |
[6] Thomas, J et al.. (2010) Toenail onychomycosis: an important global disease burden. J Clin Pharm Ther 35, 497–519.
| Toenail onychomycosis: an important global disease burden.Crossref | GoogleScholarGoogle Scholar | 20831675PubMed |
[7] Bongomin, F et al.. (2018) A review of onychomycosis due to Aspergillus species. Mycopathologia 183, 485–493.
| A review of onychomycosis due to Aspergillus species.Crossref | GoogleScholarGoogle Scholar | 29147866PubMed |
[8] Ellis, DH et al.. (1997) Non-dermatophytes in onychomycosis of the toenails. Br J Dermatol 136, 490–493.
| Non-dermatophytes in onychomycosis of the toenails.Crossref | GoogleScholarGoogle Scholar | 9155945PubMed |
[9] Shemer, A et al.. (2009) New criteria for the laboratory diagnosis of nondermatophyte moulds in onychomycosis. Br J Dermatol 160, 37–39.
| New criteria for the laboratory diagnosis of nondermatophyte moulds in onychomycosis.Crossref | GoogleScholarGoogle Scholar | 18764841PubMed |
[10] Pharmaceutical Benefits Scheme (PBS) (2021) General schedule, antifungals for systemic use, Terbinafine. Available at https://www.pbs.gov.au/medicine/item/2285G-2804N-4011D [Accessed 23 December 2021]
[11] Weitzman, I and Summerbell, RC (1995) The dermatophytes. Clin Microbiol Rev 8, 240–259.
| The dermatophytes.Crossref | GoogleScholarGoogle Scholar | 7621400PubMed |
[12] Weinberg, JM et al.. (2003) Comparison of diagnostic methods in the evaluation of onychomycosis. J Am Acad Dermatol 49, 193–197.
| Comparison of diagnostic methods in the evaluation of onychomycosis.Crossref | GoogleScholarGoogle Scholar | 12894064PubMed |
[13] Haghani, I et al.. (2013) Comparison of diagnostic methods in the evaluation of onychomycosis. Mycopathologia 175, 315–321.
| Comparison of diagnostic methods in the evaluation of onychomycosis.Crossref | GoogleScholarGoogle Scholar | 23371413PubMed |
[14] Public Health England (2016) UK Standards for Microbiology Investigations. SMI B39. Investigation of Dermatological Specimens for Superficial Mycoses. Available at https://www.gov.uk/government/publications/smi-b-39-investigation-of-dermatological-specimens-for-superficial-mycoses [Accessed 13 December 2021]
[15] Ghannoum, M et al.. (2018) Examining the importance of laboratory and diagnostic testing when treating and diagnosing onychomycosis. Int J Dermatol 57, 131–138.
| Examining the importance of laboratory and diagnostic testing when treating and diagnosing onychomycosis.Crossref | GoogleScholarGoogle Scholar | 28653769PubMed |
[16] Bruker Daltonics GmbH & Co. KG. (2021) MBT Filamentous Fungi Library 4.0 Release Notes. Revision D Doc. no. 5027071.
[17] L’Ollivier, C et al.. (2013) A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory. Med Mycol 51, 713–720.
| A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory.Crossref | GoogleScholarGoogle Scholar | 23611419PubMed |
[18] Shaw, D (2021) Matrix-assisted laser desorption/ionisation-time of flight mass spectrometry: protocol standardisation, comparison and database expansion for faster and reliable identification of dermatophytes. Mycoses 64, 926–935.
| Matrix-assisted laser desorption/ionisation-time of flight mass spectrometry: protocol standardisation, comparison and database expansion for faster and reliable identification of dermatophytes.Crossref | GoogleScholarGoogle Scholar | 33851439PubMed |
[19] Normand, AC et al.. (2018) Nucleotide sequence database comparison for routine dermatophyte identification by internal transcribed spacer 2 genetic region DNA barcoding. J Clin Microbiol 56, e00046–18.
| Nucleotide sequence database comparison for routine dermatophyte identification by internal transcribed spacer 2 genetic region DNA barcoding.Crossref | GoogleScholarGoogle Scholar | 29491019PubMed |
[20] Arabatzis, M et al.. (2007) Diagnosis of common dermatophyte infections by a novel multiplex real-time polymerase chain reaction detection/identification scheme. Br J Dermatol 157, 681–689.
| Diagnosis of common dermatophyte infections by a novel multiplex real-time polymerase chain reaction detection/identification scheme.Crossref | GoogleScholarGoogle Scholar | 17672875PubMed |
[21] Ross, IL et al.. (2020) Detection and identification of dermatophyte fungi in clinical samples using a commercial multiplex tandem PCR assay. Pathology 52, 473–477.
| Detection and identification of dermatophyte fungi in clinical samples using a commercial multiplex tandem PCR assay.Crossref | GoogleScholarGoogle Scholar | 32307094PubMed |
[22] Lin, BB et al.. (2021) Multiplex RT-PCR provides improved diagnosis of skin and nail dermatophyte infections compared to microscopy and culture: a laboratory study and review of the literature. Diagn Microbiol Infect Dis 101, 115413.
| Multiplex RT-PCR provides improved diagnosis of skin and nail dermatophyte infections compared to microscopy and culture: a laboratory study and review of the literature.Crossref | GoogleScholarGoogle Scholar | 34256251PubMed |
[23] Kabtani, J et al.. (2021) Real-time PCR assay for the detection of dermatophytes: comparison between an in-house method and a commercial kit for the diagnosis of dermatophytoses in patients from Dakar, Senegal. J Fungi (Basel) 7, 949.
| Real-time PCR assay for the detection of dermatophytes: comparison between an in-house method and a commercial kit for the diagnosis of dermatophytoses in patients from Dakar, Senegal.Crossref | GoogleScholarGoogle Scholar |
[24] Cuchí-Burgos, E et al.. (2021) Commercial real time PCR implementation for rapid diagnosis of onychomycosis: a new workflow in a clinical laboratory. Enferm Infecc Microbiol Clin (Engl Ed) 39, 326–329.
| Commercial real time PCR implementation for rapid diagnosis of onychomycosis: a new workflow in a clinical laboratory.Crossref | GoogleScholarGoogle Scholar | 34353508PubMed |
[25] Spiliopoulou, A et al.. (2015) Evaluation of a commercial PCR test for the diagnosis of dermatophyte nail infections. J Med Microbiol 64, 25–31.
| Evaluation of a commercial PCR test for the diagnosis of dermatophyte nail infections.Crossref | GoogleScholarGoogle Scholar | 25418736PubMed |
[26] Hayette, MP et al.. (2019) Clinical evaluation of the DermaGenius® Nail real-time PCR assay for the detection of dermatophytes and Candida albicans in nails. Med Mycol 57, 277–283.
| Clinical evaluation of the DermaGenius® Nail real-time PCR assay for the detection of dermatophytes and Candida albicans in nails.Crossref | GoogleScholarGoogle Scholar | 29762721PubMed |
[27] Laroche, L et al.. (2021) Epidemiological analysis of nail aspergillosis and non-dermatophyte moulds. Criteria for the involvement of Aspergillus in 102 cases of onychomycosis at Montpellier University Hospital, France (1991–2019). Mycoses 64, 1546–1553.
| Epidemiological analysis of nail aspergillosis and non-dermatophyte moulds. Criteria for the involvement of Aspergillus in 102 cases of onychomycosis at Montpellier University Hospital, France (1991–2019).Crossref | GoogleScholarGoogle Scholar | 34467565PubMed |
[28] Moreno, G and Arenas, R (2010) Other fungi causing onychomycosis. Clin Dermatol 28, 160–163.
| Other fungi causing onychomycosis.Crossref | GoogleScholarGoogle Scholar | 20347658PubMed |
[29] Singh, A et al.. (2021) Evaluation of DermaGenius® resistance real-time polymerase chain reaction for rapid detection of terbinafine-resistant Trichophyton species. Mycoses 64, 721–726.
| Evaluation of DermaGenius® resistance real-time polymerase chain reaction for rapid detection of terbinafine-resistant Trichophyton species.Crossref | GoogleScholarGoogle Scholar | 33760310PubMed |
[30] The European Committee on Antimicrobial Susceptibility Testing (2020) Overview of antifungal ECOFFs and clinical breakpoints for yeasts, moulds and dermatophytes using the EUCAST E.Def 7.3, E.Def 9.3 and E.Def 11.0 procedures. Version 2. Available at http://www.eucast.org