Correct interpretation of actinomycete imagery using scanning electron microscopy
D. İpek Kurtböke A *A Centre for Bioinnovation, School of Science, Technology and Engineering, University of the Sunshine Coast, Maroochydore DC, Qld 4558, Australia.

Dr Kurtböke is currently a senior lecturer at the University of the Sunshine Coast (USC) in Australia and one of the members of the Genecology Research Centre of the USC, conducting research in applied, industrial and environmental microbiology. She is an internationally reputed actinomycetologist and she has been in the field of biodiscovery since 1982 conducting research into discovery of novel and potent therapeutic compounds produced by actinomycetes in Turkey, Italy, the UK, and Australia with leading pharmaceutical companies. She has been an Executive Board member of the World Federation of Culture Collections (WFCC) since 2000, currently serving her second term as the President of the Federation. She is also one of the members of the International Committee on Taxonomy of Viruses (ICTV)’s, Bacterial Viruses Subcommittee. She has editorial duties in different journals including Marine Drugs, Diversity and Frontiers Marine Science/Marine Biotechnology. |
Microbiology Australia 43(1) 28-31 https://doi.org/10.1071/MA22009
Submitted: 17 January 2022 Accepted: 1 March 2022 Published: 22 April 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Antibiotic discovery was one of the most significant advances in therapeutic medicine following the advances in fermentation technology owing to Howard Florey and his associates. The ‘Golden era’ of antibiotics following the first discoveries in the laboratory of Waksman and his colleagues from a group of microorganisms known as actinomycetes lasted for 34 years (1940–1974). These fascinating microorganisms especially the members of the genus Streptomyces gave us the majority of the known antibiotics we use today, like streptomycin, kanamycin, neomycin, gentamicin, vancomycin and many more. To be able to produce these antibiotics in large-scale, the producer actinomycetes had to be selectively isolated. This resulted in a collaboration of over 40 laboratories from 18 different countries called ‘The International Streptomyces Project (ISP)’. The isolates generated in this project were studied in-depth including their morphologies together with their bioactivity. One of the components of these investigation was the correct interpretation of actinomycete morphology including the use of scanning electron microscopy. At the end of the first European Actinomycete Conference in Bradford University in England (1984), I had the opportunity to be trained by late Professor Cross on actinomycete growth morphologies. Thirty-eight years later when I witness the frequent difficulties students encounter in the interpretation of the actinomycete SEM images, I decided to write this paper and pass the skills given to me by late Professor Cross to the younger generation.
Keywords: Actinomycetales, Actinomycetes, Actinomycetia, Actinomycetota, growth morphology, SEM imagery, Streptomyces, taxonomy.
In Memoriam, Professor Tom Cross, University of Bradford, UK.
Current classification of actinomycetes
First discovery of a species within this group of bacteria dates to Harz in 18771 by the description of Actinomyces bovis. Later Buchanan named the order Actinomycetales2 and in the subsequent year named the family Actinomycetecaea.3 Stackebrandt et al.4 proposed a new Class called Actinobacteria under the Domain Bacteria, followed by the creation of the phylum with the same name.5,6 In this restructure order, Actinomycetales was confined to the original Hartz cluster that is comprised of non-mycelial taxa (e.g. Actinomyces).
The use of the same name both for the phylum and the class was, however, not preferable, and recently a new class named Actinomycetia was proposed by Salam et al.7 covering all members of the former order Actinomycetales.8 Here caution must be exercised as currently the order Actinomycetales covers the family Actinomyceteceae (Buchanan original description).9 So, families like Streptomyceteceae that formerly belonged to this order10 cannot be located under this order any longer. In addition, in 2021 the name of the phylum Actinobacteria revised again into Actinomycetota.9 In summary, the phylum Actinomycetota is the former phylum Actinobacteria. The proposed Class Actinomycetia is the former class Actinobacteria. Class Actinomycetia also covers the members of the former order Actinomycetales, which is different to the current order Actinomycetales. Finally, to conclude, every member under the class Actinomycetia is an actinomycete (actinomycetes in plural) in a general term that has been in use over 70 years.10
Another important point is that RNA oligonucleotide studies showed that the possession of branched hyphae (e.g. Thermoactinomyces) should not automatically place a bacterium within the class Actinomycetia, nor should the inability of an organism to form branching filaments (e.g. Arthrobacter, Cellulomonas and Rothia) necessarily exclude it from this taxon. Accordingly, genus Thermoactinomyces with its low guanine and cytosine content and endospore forming ability was removed from the actinomycete cluster and placed into the family Bacillaceae.11
Distinctive growth morphologies of actinomycetes
Whichever family they belong to, actinomycetes only display four different types of growth morphologies (Table 1). Full understanding of these structures is imperative during interpretation of the SEM images.
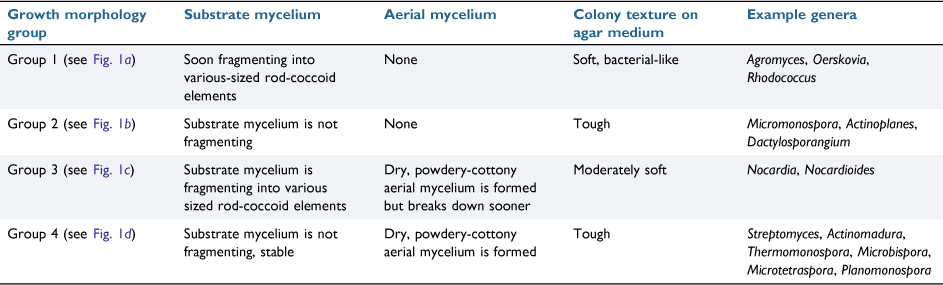
|
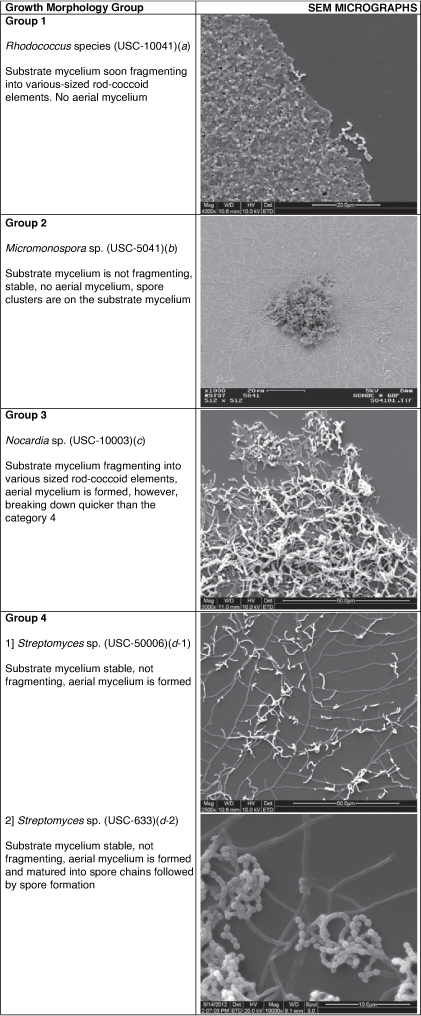
|
It is also important that intact material is not damaged during processing for the SEM such as when gradual dehydration in alcohol is done prior to critical point drying. Any damage during processing can result in wrong interpretation of the image e.g. spore surface morphology, especially for the structures ‘rugose’ and ‘warty’.8,14
Another important aspect is the timing of the examination for growth morphologies. Different microscope slide preparations should be prepared and examined in consecutive days. Young colonies will exhibit different morphologies as full maturation is not complete as well as too old colonies will result in collapse of spore chains. Spore surfaces also should be examined at right times (this differs for all above listed four different growth morphologies) to be able to identify spore surface structure correctly. As an example, a smooth looking spore surface might change into a ‘hairy’ or ‘spiny’ structure8 later in the growth cycle (Fig. 2).
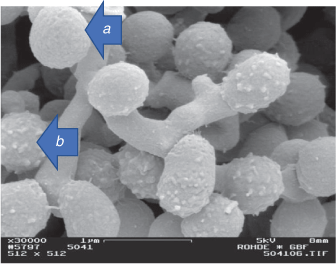
|
As noted by the late Professor Cross (1989),15 choice of media is important as most sporoactinomycetes would require special media to allow differentiation and development of characteristic spores and pigments. His examples include, the transformation of pale, shiny, hard colonies of a Streptomyces species on nutrient agar into bright yellow colonies with a powdery white aerial mycelium and spirals of arthrospores when the organism is subcultured onto a more suitable growth medium such as oatmeal or inorganic salt starch agars.16 More recent examples of such differences in growth can be seen in recent publications of English et al.17 and Kurtböke.18
Again, the late Professor Cross (1989)15 highlighted that the actinomycete outgrowths starts from fragments of mycelium and develop into hyphae that penetrate the agar forming the substrate mycelium and hyphae that branch repeatedly and become cemented together on the surface of the agar to form a tough, leathery colony. He also added that actinomycete growth can be slow, a branching mycelium growing at the surface of transparent agar can be seen with the aid of a microscope after 24 h, and visible colonies may appear in 3–4 days, but mature aerial mycelium with spores may take 7–14-days to develop, and some very slow growing strains may require up to a month of incubation. Lengthy incubation times can result in evaporation of the medium, so thick agar plates are required. Thermophilic species incubated at high temperatures require a humid incubator.
A useful diagram of the developmental life cycle of a Streptomyces species is provided by Barka et al.19 illustrating growth from sporulation to development of substrate (vegetative) and then aerial mycelium leading to septation and formation of spores.
Sample preparation for SEM
Streaking actinomycete spores/hyphal fragments on agar medium
At all times a ‘rough, stiff loop is essential for abrading the colony and collecting sufficient mycelial fragments for an efficient transfer’. Spore suspensions can also be prepared first and used for streaking.15 They can be prepared ‘by detaching the spores from aerial hyphae with a loop or scraper and placing them in a suspending medium containing a wetting agent’. The arthrospores of streptomycetes are hydrophobic because of the enveloping sheath, and the wetting agent aids their even suspension in the diluent. Free spores may also be removed from lawns of aerial mycelium by rolling glass beads or agar cylinders over the surface.15
Inclined cover slip technique
Best results can be achieved using inclined glass coverslip technique15 and Oatmeal agar supplemented with yeast extract.12 Once the actinomycete is streaked plated onto this medium, round cover slips can be embedded onto the streaked lines with a 45° angle. Growth starts and developing mycelia simultaneously moves onto the coverslip. Multiple cover slips would allow replication and observations to be conducted at different growth times.
Once the growth is sufficient the cover slip can be removed, place onto SEM stubs, fixed with osmium tetroxide, gradually dehydrated in alcohol before being subjected to critical point drying and subsequently coating with gold.20,21 In this final stage, it is imperative that experienced technical officers who have acquired knowledge on the use of SEM assist the researcher who is engaged in image interpretation.
Modified techniques were also developed, such as the one by Prakash and Nawani22 in which lyophilisation is used rather than chemical fixatives and dehydrating agents.
Conclusions
Again, as stated by the late Professor Cross,15 ‘one requires patience when working with actinomycetes, and the ability to plan and run several experiments concurrently to avoid wasting time’. I would like to add the importance of students understanding the long and arduous route from being a novice to becoming an expert. Hard work and perseverance is important to patiently build their knowledge and gain laboratory skills.23 What is presented here is the outcome of 40 years of continuous learning and skill building in the field of actinomycetology.24 Understanding the value of team work as well as appreciating the roles of other disciplines and experts such as technical staff members in EM units, without whom the quality micrographs cannot be produced, is also imperative for novices.
Data availability
Data is embedded in the text as SEM micrographs.
Conflicts of interest
The author declares no conflicts of interest.
Declaration of funding
This research did not receive any specific funding.
Acknowledgements
I thank Professor Manfred Rohde, GBF (Figs 1b, 2), Germany, and Mrs Rachel Hancock, Central Analytical Research Facility operated by the Institute for Future Environments, Queensland, University of Technology (QUT), Brisbane, Australia (Figs 1a, c, d, 2), for their invaluable assistance with the SEM imagery.
References
[1] Harz, CO et al.. (1879) Actinomyces bovis ein neuer Schimmel in dem Geweben des rindes. Jahrebericht der K Central-Thieraznei Schule in Müchen für 1877–1879 5, 125–140.[2] Buchanan, RE et al.. (1917) Studies in the nomenclature and classification of the bacteria II. The primary subdivisions of the schizomycetes. J Bacteriol 2, 155–164.
| Studies in the nomenclature and classification of the bacteria II. The primary subdivisions of the schizomycetes.Crossref | GoogleScholarGoogle Scholar | 16558735PubMed |
[3] Buchanan, RE et al.. (1918) Studies in the classification and nomenclature of the bacteria VIII. The subgroups and genera of the Actinomycetales. J Bacteriol 3, 403–406.
| Studies in the classification and nomenclature of the bacteria VIII. The subgroups and genera of the Actinomycetales.Crossref | GoogleScholarGoogle Scholar | 16558803PubMed |
[4] Stackebrandt, E et al.. (1997) Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Evol Microbiol 47, 479–491.
| Proposal for a new hierarchic classification system, Actinobacteria classis nov.Crossref | GoogleScholarGoogle Scholar |
[5] Goodfellow M (2012) Phylum XXVI. Actinobacteria phyl. nov. In Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 5, The Actinobacteria, Part A. (Goodfellow M, et al., eds). pp. 33–34. Springer, New York, NY.
[6] Nouioui, I et al.. (2018) Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol 9, 2007.
| Genome-based taxonomic classification of the phylum Actinobacteria.Crossref | GoogleScholarGoogle Scholar | 30186281PubMed |
[7] Salam, N et al.. (2020) Update on the classification of higher ranks in the phylum Actinobacteria. Int J Syst Evol Microbiol 70, 1331–1355.
| Update on the classification of higher ranks in the phylum Actinobacteria.Crossref | GoogleScholarGoogle Scholar | 31808738PubMed |
[8] Williams ST et al. (eds) (1989) Bergey’s Manual of Systematic Bacteriology, vol. 4. Williams and Wilkins, Baltimore.
[9] Oren, A and Garrity, GM (2021) Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol 71, 005056.
| Valid publication of the names of forty-two phyla of prokaryotes.Crossref | GoogleScholarGoogle Scholar |
[10] Waksman, SA and Henrici, AT (1943) The nomenclature and classification of the actinomycetes. J Bacteriol 46, 337–341.
| The nomenclature and classification of the actinomycetes.Crossref | GoogleScholarGoogle Scholar | 16560709PubMed |
[11] Park, YH et al.. (1993) Suprageneric classification of Thermoactinomyces vulgaris by nucleotide sequencing of 5S ribosomal. RNA Zentralblatt für Bakteriologie 278, 469–478.
| Suprageneric classification of Thermoactinomyces vulgaris by nucleotide sequencing of 5S ribosomal.Crossref | GoogleScholarGoogle Scholar | 7688999PubMed |
[12] Williams ST, Wellington EMH (1982) Actinomycetes. In Methods of Soil Analysis, Part 2. Chemical, Microbiological Properties, 2nd edn (Page AL et al., eds). pp. 969–987.
[13] Goodfellow M, Cross T (1984) Classification. In The Biology of the Actinomycetes (Goodfellow M, et al., eds). pp. 7–164. Academic Press, London.
[14] Locci R (1989) Streptomycetes and related genera, Section 29. In Bergey’s Manual of Systematic Bacteriology, vol. 4, (Williams ST et al., eds). pp. 2451–2492. Williams and Wilkins, Baltimore.
[15] Cross T (1989) Growth and examination of actinomycetes-some guidelines. In Bergey’s Manual of Systematic Bacteriology, vol. 4 (Williams ST et al., eds). pp. 2340–2343. Williams and Wilkins, Baltimore.
[16] Nonomura, H et al.. (1974) Key for classification and identification of 458 species of the streptomycetes included in the ISP. J Ferment Technol 52, 78–92.
[17] English, AL et al.. (2017) Evaluation of fermentation conditions triggering increased antibacterial activity from a near-shore marine intertidal environment-associated Streptomyces species. Synth Syst Biotechnol 2, 28–38.
| Evaluation of fermentation conditions triggering increased antibacterial activity from a near-shore marine intertidal environment-associated Streptomyces species.Crossref | GoogleScholarGoogle Scholar | 29062959PubMed |
[18] Kurtböke DI (2017) Bioactive actinomycetes: reaching rarity through sound understanding of selective culture and molecular diversity. In Microbial Resources: From Functional Existence in Nature to Applications (Kurtböke DI, ed.). pp. 45–76. Academic Press.
[19] Barka, EA et al.. (2016) Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev 80, 1–43.
| Taxonomy, physiology, and natural products of Actinobacteria.Crossref | GoogleScholarGoogle Scholar | 26609051PubMed |
[20] Williams, ST et al.. (1970) Further investigations of actinomycetes by scanning electron microscopy. Microbiology 62, 67–73.
| Further investigations of actinomycetes by scanning electron microscopy.Crossref | GoogleScholarGoogle Scholar |
[21] Williams, ST and Davies, FL (1967) Use of a scanning electron microscope for the examination of actinomycetes. Microbiology 48, 171–177.
| Use of a scanning electron microscope for the examination of actinomycetes.Crossref | GoogleScholarGoogle Scholar |
[22] Prakash, D and Nawani, NN (2014) A rapid and improved technique for scanning electron microscopy of actinomycetes. J Microbiol Methods 99, 54–57.
| A rapid and improved technique for scanning electron microscopy of actinomycetes.Crossref | GoogleScholarGoogle Scholar | 24556287PubMed |
[23] Kurtböke DI (ed.) (2022) Importance of Microbiology Teaching and Microbial Resource Management for Sustainable Futures. Academic Press.
[24] Kurtböke, I et al.. (2012) From Actinomycin onwards: Actinomycete success stories. Microbiol Aust 33, 108–110.
| From Actinomycin onwards: Actinomycete success stories.Crossref | GoogleScholarGoogle Scholar |


