Mitigating greenhouse gas emissions from waste treatment through microbiological innovation
Gaofeng Ni A *A Department of Microbiology, Biomedicine Discovery Institute, Monash University, Clayton, Vic., Australia.

Dr Gaofeng Ni is a Postdoctoral Research Fellow and Industrial Microbiology Team Leader at the One Health Microbiology Laboratory led by Professor Chris Greening at Monash University. His expertise ranges from biotechnological innovation in industrial wastewater treatment to the molecular physiology and metabolism of microorganisms living at extreme conditions. |
Microbiology Australia 44(1) 22-26 https://doi.org/10.1071/MA23006
Submitted: 26 January 2023 Accepted: 2 February 2023 Published: 1 March 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
The emission of greenhouse gases (GHGs) from the treatment of municipal, agricultural and industrial waste occurs in virtually every city on our planet. This is due to various microbial activities at different stages of waste treatment. Traditional treatment methods have a significant environmental impact, producing methane, carbon dioxide and nitrous oxide emissions, in addition to demanding high energy input and having low treatment efficiencies. To address these issues, the Australian water and waste sectors are shifting towards the adoption of next-generation, carbon-neutral treatment options. Here I discuss our current knowledge gaps in mitigating GHG emissions from waste streams, with a focus on wastewater treatment plants. I highlight the application of real-time genomics to identify sources of GHG emissions, monitor mitigation efforts, assist process operation and guide plant operations. I also emphasise recent innovations of microbial processes that capture GHG from waste and upgrade them into higher value products. Ultimately, combined effort across disciplines is required to proactively mitigate the global threat of climate change.
Greenhouse gas emissions from waste streams
The first systematic quantification of methane and carbon dioxide emissions from wastewater treatment plants (WWTPs) dates back to 1993, at a full-scale wastewater treatment plant serving 12 500 inhabitants in Durham, NH, USA.1 Aeration is an essential part of modern wastewater treatment, which provides oxygen to support the respiration of aerobic microorganisms to degrade organic carbon and nitrogen compounds. The activated sludge process (Fig. 1), which uses microbial flocs or granules to remove pollutants such as carbon, nitrogen and phosphorus from wastewater, depends on this aeration.2 This is a crucial process to safeguard public and environmental health,2 but it also leads to the emission of carbon dioxide through the respiration of heterotrophic microorganisms and nitrous oxide (N2O) through activities of nitrifying and denitrifying microorganisms. Anaerobic digestion (Fig. 1) is the biological treatment of waste sludge in the absence of oxygen to stabilise organic matter while producing biogas, containing methane and carbon dioxide.3 Anaerobic digestion has become a mature technology widely applied in WWTPs in Australia and across the world. Disposal and land application of digested sludge also results in carbon dioxide emissions. It is estimated that methane emission from wastewater treatment and landfills (Fig. 1) accounted for 21% of global methane emissions in 2021.4 Established sewage networks and wastewater treatment facilities in nearly every city make WWTPs among the largest point sources of GHG emissions and also ideal entry points to mitigate GHG emissions (Fig. 1).
Pinpointing sources and regulators
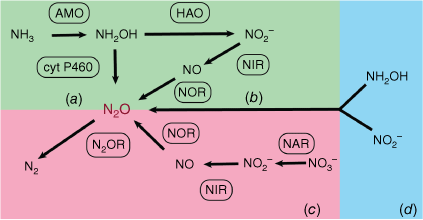
Microorganisms in wastewater treatment play key roles in regulating GHG emissions, so it is critical to understand their physiology and ecology to effectively reduce these emissions. The respiration of microorganisms in standard treatment technologies, such as the anaerobic–anoxic–oxic (AAO) process or sequencing batch reactors (SBR), leads to direct carbon dioxide discharge into the atmosphere. Anaerobic digestion is the major source of methane emission based on estimation from eight Austrian WWTPs5; and it also produces carbon dioxide to a less extent.6 More recently, it has been recognised that much nitrous oxide is being released from WWTPs at significant rates and can contribute up to 83% of the emissions footprint of WWTPs expressed as CO2 equivalents.7 Both nitrifying and denitrifying microorganisms mediate the release and consumption of nitrous oxide through four known processes: (a) as a by-product of hydroxylamine (NH2OH) oxidation by nitrifiers; (b) through nitrifier denitrification during the reduction of nitric oxide (NO); (c) denitrification by heterotrophic microorganisms; and (d) codenitrification, where one nitrogen atom in N2O originates from hydroxylamine and the other from nitrite8,9 (Fig. 2). Compared to carbon dioxide and methane, we are still in the preliminary stage of understanding nitrous oxide regulation pathways, and require integrated research efforts of the microbiology, biochemistry and biogeochemistry underlining these processes.
|
We currently lack microbiology-based prediction tools for GHG emissions. Isotopic techniques have been developed to quantify the contribution of nitrous oxide production and consumption from different regulatory pathways, but they are labour-intensive and hindered by the presence of unidentified microbial pathways.9 Additionally, these methods require elaborate laboratory procedures and equipment, making them difficult to operate remotely. The mobile tracer gas dispersion method can estimate major sources of GHG emissions on a plant level based on the dispersion patterns of methane and nitrous oxide in the atmosphere,10 but it does not allow for long-term and continuous monitoring and is heavily dependent on stable wind patterns.11 By contrast, process unit quantification using the standard floating hood technique has methodological limitations in addressing spatial variability of GHG emissions or inaccuracies caused by mass transfer alterations inside the hood, as well as practical limitations in terms of difficulties in being deployed at foaming and turbulent wastewaters or treatment units with obstacles such as surface aerators.11 These limitations make it challenging in extrapolating GHG emission estimation from floating hood measurements to larger areas and capturing the spatial dynamics of GHG emissions at the plant level.11 The main drawback of these methods is that they fail to uncover the microbial regulatory processes, lack the ability to consider metabolic diversity, and are not able to predict greenhouse gas emissions with optimal precision on a spatial and temporal scale. These models rely on generic pathways to describe overall nitrous oxide emission; they do not address the inner workings of microbial communities; and therefore, cannot discern the contributions of individual pathways as described above (Fig. 2).
Predictive understanding of GHG emissions from the waste sector depends on better understanding the microbial mediators and mitigators of this process. This requires systematic investigation of the physiology of GHG cycling microorganisms, including methanogens, nitrifiers, denitrifiers and heterotrophic bacteria, found in all locations of wastewater treatment plants (WWTPs) known for GHG emissions, including aeration tanks of the AAO process or SBR reactors, anaerobic digesters, sludge drying lagoons, clarifiers and disinfection units.12,13 Culturing is the definitive way to characterise the capacity of these microorganisms to produce or consume GHGs and understand how this cycling varies depending on environmental conditions. Rapid advancements in sequencing technologies have significantly decreased the cost of sequencing, with increased throughput and higher accuracy using less genetic material. The use of portable sequencers, such as the Oxford nanopore minion (ONION), coupled with cloud computing, has allowed specialists to observe shifts in methanogenic communities on-site at WWTP operations in near real time.14 This can provide early warning of microorganisms that have potential to increase GHG emissions, therefore inform mitigation strategies such as pH alteration to suppress the growth of these organisms. By integrating physiological data of these microorganisms with real-time genomics techniques, biogeochemical measurements and supervised machine-learning approaches, it will be increasingly possible to develop predictive models for GHG emissions across space and time. However, this requires further calibration based on operational data such as temperature, dissolved oxygen and nutrient load.
Using microbiology to reduce and recycle emissions
Real-time genomics to guiding treatment operations
There are several options to use real-time genomics to guide GHG mitigation and plant operation. Nitrous oxide reductase (N2OR, Fig. 2) is the only known enzyme responsible for reducing nitrous oxide emissions.15 By metagenomics and metatranscriptomics, the activity of microorganisms expressing NosZ can be estimated at hotspots of GHG emissions through the identification of microorganisms encoding the NosZ gene and their mRNA transcripts. This information can be used to identify optimal operational conditions (such as carbon and nitrogen load, pH, dissolved oxygen level, and temperature) at which N2OR activities are the highest in real time. Additionally, monitoring the expression of cytochrome P460 or nitric oxide reductase (cyt P460 and NOR, Fig. 2) at different treatment conditions can reveal metabolic dynamics of nitrous oxide emission, which can guide process operation to reduce GHG emissions.
Moreover, real-time genomics can assist optimisation of treatment processes. Traditional treatment methods have drawbacks such as high energy consumption, the need for a carbon source, fugitive GHG emissions, low energy recovery efficiency and excessive sludge production.16 Recent advances in combining anaerobic methane-oxidising archaea and bacteria with existing treatment techniques have the potential to overcome these barriers by utilising methane as a carbon and energy source to enhance nitrogen removal.17 However, one limitation of using these anaerobic microorganisms is their slow growth (doubling time >10 days); it can take longer than a year before performance improvement is visible.16 Genomic sequencing and real-time analysis can greatly aid in optimising reactor configuration and operational conditions by providing immediate feedback on the ecological (such as abundance) and physiological states (expression of genes related to growth, adaptation and stress response) of these anaerobic methanotrophs.
Furthermore, real-time genomics can provide early warnings of unwanted microorganisms. The stable performance of wastewater treatment processes relies on the healthy composition of microbial communities. For instance, the notoriously long-standing problem of poor sludge settleability in clarifiers (also known as ‘sludge bulking’) is caused by filamentous bacteria.18 Partial nitritation coupled with the anammox process (anaerobic ammonia oxidation) is a highly promising treatment method as it reduces aeration costs by up to 60% aeration costs and eliminates the need for organic carbon dosing.19,20 However, the appearance of nitrite oxidising bacteria, which competes with anammox bacteria for the substrate nitrite, can impede the process. The emergence of these unwanted microorganisms can deteriorate the treatment processes, but they can only be detected when they have already established themselves. This leaves treatment specialists in a reactive position, as current methods such as physiochemical measurements cannot detect them early. Real-time genomics can predict the emergence of these unwanted microorganisms through early detection by genome-resolved metagenomics or marker gene-based sequencing combined with physiological data. This enables targeted response strategies, such as lowering dissolved oxygen and increasing ammonia concentration to suppress nitrite oxidising bacteria.21 These genomics solutions require refinement and testing for improved robustness, increased accuracy and reduced cost before full-scale implementation.
Converting methane to value-added carbon
One promising way to mitigate GHG emissions from the waste sector is to convert methane produced by methanogens into value-added carbon. By using the metabolic capacities of methanotrophic microorganisms,22 it is possible to convert methane into higher-value products such as methanol, polyhydroxyalkanoates (PHA), biopolymers and single-cell proteins (SCPs).23 The first step in these conversions is methane oxidation to methanol catalysed by the soluble form of methane monooxygenase (sMMO) or its particulate form (pMMO) (Eqn 1).24
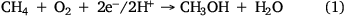
where ΔG0 = –111 kJ mol–1.
The most well-known aerobic methanotrophs for biotechnological exploration include Methylococcus capsulatus Bath, Methylomonas spp. and Methylosinus trichosporium OB3b. However, aerobic conversion requires an oxygen input of at least a 1:1 methane to oxygen ratio. Although oxygen is widely available, it costs from US$98.4 to $123.0 per tonne depending on the cost of energy,25 which increases the total cost.26 Anaerobic methanotrophic (ANME) archaea and ‘Candidatus Methylomirabilis’ (NC10) bacteria are promising candidates for methane biorefinery through anaerobic or intra-aerobic oxidation,27 with significant research and development to understand their metabolism and ecology.
Innovative CO2 capture
By 2050, microbial proteins are estimated to replace between 10 and 19% of conventional crop-based animal feed protein demand, depending on socio-economic development and microbial protein production from GHGs.28 This can substantially decrease global cropland expansion, GHG emission and nitrogen pollution. Carbon dioxide produced at WWTPs is conventionally perceived as waste carbon, but it can be an ideal stock feed for a variety of novel carbon dioxide capture technologies because it is produced at a point source. In addition, aerobic hydrogen oxidising microorganisms such as Cupriavidus necator are considered to be ‘powerful microbial actuators’ due to their ability to use hydrogen to conserve energy (Eqn 2) and fix carbon dioxide into cellular material,29 producing single-cell proteins with increased value and high yield.
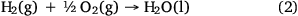
where ΔG0 = –237.1 kJ mol–1.
Examples include Sulfuricurvum that was shown to provide stable performance over nearly 100 days in a laboratory scale study, converting carbon dioxide, ammonium and hydrogen into high quality microbial proteins.30 Also, purple phototrophic bacteria are anoxygenic phototrophs capable of using a variety of organic (volatile fatty acids) and inorganic (hydrogen gas, hydrogen sulfide and ferrous iron) substrates for anaerobic photoheterotrophic or photoautotrophic growth.31 Their unique capacity for near-infrared light absorption by bacteriochlorophylls or visible light absorption by carotenoids highlights their suitability for carbon dioxide capture and single-cell protein production in Australia, where solar radiation is abundant.32 Additionally, carbon dioxide capturing through microalgae to produce biodiesel offers another interesting avenue to harvest solar energy to fix carbon dioxide with high biomass yield.33 Finally, it is also possible to produce biochemicals such as ethanol, methane, hydrogen and propanediol through microbial electrosynthesis with renewable electricity as input.34
Conclusion
Climate change poses an imminent existential threat to humanity. Innovative microbiological technologies such as real-time genomics can provide crucial information to mitigate GHG emission at treatment plants. Microbiology also offers a range of solutions for capturing and valorising GHGs (Fig. 1). As microbiologists, we strive to reframe the current paradigm of intensive resource exploration and waste discharge into valorisation of waste and GHG mitigation with improved treatment processes. To make this happen, we need combined expertise in molecular and structural microbiology, process engineering, mathematical modelling. We also need to foster collaboration among waste management, academic research, and renewable energy sectors to limit global temperature increase to below 1.5°C in the next two decades.35
Data availability
This research article does not contain any data. All information presented in this article is based on previously published research or other sources, and no original data were collected or analysed.
Conflicts of interest
The author declares that they have no conflicts of interest.
Declaration of funding
This work is supported by a Monash University Early Career Research Postdoctoral Fellowship (ECPF23-8566329039) awarded to the author.
Acknowledgements
The author gratefully acknowledges Prof. Chris Greening at Monash University (Australia) for constructive discussions and feedback to this work.
References
[1] Czepiel, PM et al. (1993) Methane emissions from municipal wastewater treatment processes. Environ Sci Technol 27, 2472–2477.| Methane emissions from municipal wastewater treatment processes.Crossref | GoogleScholarGoogle Scholar |
[2] Wu, L et al. (2019) Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat Microbiol 4, 1183–1195.
| Global diversity and biogeography of bacterial communities in wastewater treatment plants.Crossref | GoogleScholarGoogle Scholar |
[3] Mata-Alvarez, J et al. (2014) A critical review on anaerobic co-digestion achievements between 2010 and 2013. Renew Sustain Energy Rev 36, 412–427.
| A critical review on anaerobic co-digestion achievements between 2010 and 2013.Crossref | GoogleScholarGoogle Scholar |
[4] Olivier JGJ (2022) Trends in global CO and total greenhouse gas emissions: 2021 Summary Report. PBL Netherlands Environmental Assessment Agency, The Hague, Netherlands. https://www.pbl.nl/sites/default/files/downloads/pbl-2022-trends-in-global-co2-and_total-greenhouse-gas-emissions-2021-summary-report_4758.pdf
[5] Parravicini, V et al. (2016) Greenhouse gas emissions from wastewater treatment plants. Energy Procedia 97, 246–253.
| Greenhouse gas emissions from wastewater treatment plants.Crossref | GoogleScholarGoogle Scholar |
[6] Zhang, J et al. (2021) Novel anaerobic digestion and carbon dioxide emissions efficiency analysis of food waste treatment based on SBM-DEA model. J Clean Prod 328, 129591.
| Novel anaerobic digestion and carbon dioxide emissions efficiency analysis of food waste treatment based on SBM-DEA model.Crossref | GoogleScholarGoogle Scholar |
[7] Daelman, MRJ et al. (2013) Methane and nitrous oxide emissions from municipal wastewater treatment – results from a long-term study. Water Sci Technol 67, 2350–2355.
| Methane and nitrous oxide emissions from municipal wastewater treatment – results from a long-term study.Crossref | GoogleScholarGoogle Scholar |
[8] Müller, C et al. (2014) Quantification of N2O emission pathways via a 15N tracing model. Soil Biol Biochem 72, 44–54.
| Quantification of N2O emission pathways via a 15N tracing model.Crossref | GoogleScholarGoogle Scholar |
[9] Duan, H et al. (2017) Quantifying nitrous oxide production pathways in wastewater treatment systems using isotope technology – a critical review. Water Res 122, 96–113.
| Quantifying nitrous oxide production pathways in wastewater treatment systems using isotope technology – a critical review.Crossref | GoogleScholarGoogle Scholar |
[10] Delre, A et al. (2018) Emission quantification using the tracer gas dispersion method: the influence of instrument, tracer gas species and source simulation. Sci Total Environ 634, 59–66.
| Emission quantification using the tracer gas dispersion method: the influence of instrument, tracer gas species and source simulation.Crossref | GoogleScholarGoogle Scholar |
[11] Parravicini V et al. (2022) Full-scale quantification of N2O and CH4 emissions from urban water systems. In Quantification and Modelling of Fugitive Greenhouse Gas Emissions from Urban Water Systems (Ye L et al., eds). pp. 91–132. IWA Publishing.
| Crossref |
[12] Caniani, D et al. (2019) CO2 and N2O from water resource recovery facilities: evaluation of emissions from biological treatment, settling, disinfection, and receiving water body. Sci Total Environ 648, 1130–1140.
| CO2 and N2O from water resource recovery facilities: evaluation of emissions from biological treatment, settling, disinfection, and receiving water body.Crossref | GoogleScholarGoogle Scholar |
[13] Ye L et al. (eds) (2022) Quantification and Modelling of Fugitive Greenhouse Gas Emissions from Urban Water Systems. IWA Publishing.
| Crossref |
[14] Hardegen, J et al. (2018) Methanogenic community shifts during the transition from sewage mono-digestion to co-digestion of grass biomass. Bioresour Technol 265, 275–281.
| Methanogenic community shifts during the transition from sewage mono-digestion to co-digestion of grass biomass.Crossref | GoogleScholarGoogle Scholar |
[15] Hu, L et al. (2022) NosZ gene cloning, reduction performance and structure of Pseudomonas citronellolis WXP-4 nitrous oxide reductase. RSC Adv 12, 2549–2557.
| NosZ gene cloning, reduction performance and structure of Pseudomonas citronellolis WXP-4 nitrous oxide reductase.Crossref | GoogleScholarGoogle Scholar |
[16] Liu, T et al. (2019) Enhancing mainstream nitrogen removal by employing nitrate/nitrite-dependent anaerobic methane oxidation processes. Crit Rev Biotechnol 39, 732–745.
| Enhancing mainstream nitrogen removal by employing nitrate/nitrite-dependent anaerobic methane oxidation processes.Crossref | GoogleScholarGoogle Scholar |
[17] Lim, ZK et al. (2021) Versatility of nitrite/nitrate-dependent anaerobic methane oxidation (n-DAMO): first demonstration with real wastewater. Water Res 194, 116912.
| Versatility of nitrite/nitrate-dependent anaerobic methane oxidation (n-DAMO): first demonstration with real wastewater.Crossref | GoogleScholarGoogle Scholar |
[18] Nittami, T and Batinovic, S (2022) Recent advances in understanding the ecology of the filamentous bacteria responsible for activated sludge bulking. Lett Appl Microbiol 75, 759–775.
| Recent advances in understanding the ecology of the filamentous bacteria responsible for activated sludge bulking.Crossref | GoogleScholarGoogle Scholar |
[19] Jetten, MSM et al. (1997) Towards a more sustainable municipal wastewater treatment system. Water Sci Technol 35, 171–180.
| Towards a more sustainable municipal wastewater treatment system.Crossref | GoogleScholarGoogle Scholar |
[20] Wang, Z et al. (2022) A 20-year journey of partial nitritation and anammox (PN/A): from sidestream toward mainstream. Environ Sci Technol 56, 7522–7531.
| A 20-year journey of partial nitritation and anammox (PN/A): from sidestream toward mainstream.Crossref | GoogleScholarGoogle Scholar |
[21] Le, L-T et al. (2020) Suppression of nitrite-oxidizing bacteria under the combined conditions of high free ammonia and low dissolved oxygen concentrations for mainstream partial nitritation. Environ Technol Innov 20, 101135.
| Suppression of nitrite-oxidizing bacteria under the combined conditions of high free ammonia and low dissolved oxygen concentrations for mainstream partial nitritation.Crossref | GoogleScholarGoogle Scholar |
[22] Jiang, H et al. (2010) Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering. Biochem Eng J 49, 277–288.
| Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering.Crossref | GoogleScholarGoogle Scholar |
[23] Pieja, AJ et al. (2017) Methane to bioproducts: the future of the bioeconomy? Curr Opin Chem Biol 41, 123–131.
| Methane to bioproducts: the future of the bioeconomy?Crossref | GoogleScholarGoogle Scholar |
[24] Khirsariya, P and Mewada, RK (2013) Single step oxidation of methane to methanol – towards better understanding. Procedia Eng 51, 409–415.
| Single step oxidation of methane to methanol – towards better understanding.Crossref | GoogleScholarGoogle Scholar |
[25] Rao P, Muller M (2007) Industrial oxygen: its generation and use. In ACEEE Summer Study on Energy Efficiency in Industry. pp. 6-124–6-135. European Council for an Energy Efficient Economy. https://www.aceee.org/files/proceedings/2007/data/papers/78_6_080.pdf
[26] Kalyuzhnaya MG (2016) Chapter 13. Methane biocatalysis: selecting the right microbe. In Biotechnology for Biofuel Production and Optimization (Eckert CA, Trinh CT, eds). pp. 353–383. Elsevier.
| Crossref |
[27] Cai, C et al. (2021) Roles and opportunities for microbial anaerobic oxidation of methane in natural and engineered systems. Energy Environ Sci 14, 4803–4830.
| Roles and opportunities for microbial anaerobic oxidation of methane in natural and engineered systems.Crossref | GoogleScholarGoogle Scholar |
[28] Pikaar, I et al. (2018) Decoupling livestock from land use through industrial feed production pathways. Environ Sci Technol 52, 7351–7359.
| Decoupling livestock from land use through industrial feed production pathways.Crossref | GoogleScholarGoogle Scholar |
[29] Khosravi-Darani, K et al. (2013) Microbial production of poly(hydroxybutyrate) from C1 carbon sources. Appl Microbiol Biotechnol 97, 1407–1424.
| Microbial production of poly(hydroxybutyrate) from C1 carbon sources.Crossref | GoogleScholarGoogle Scholar |
[30] Matassa, S et al. (2016) Autotrophic nitrogen assimilation and carbon capture for microbial protein production by a novel enrichment of hydrogen-oxidizing bacteria. Water Res 101, 137–146.
| Autotrophic nitrogen assimilation and carbon capture for microbial protein production by a novel enrichment of hydrogen-oxidizing bacteria.Crossref | GoogleScholarGoogle Scholar |
[31] Capson-Tojo, G et al. (2020) Purple phototrophic bacteria for resource recovery: challenges and opportunities. Biotechnol Adv 43, 107567.
| Purple phototrophic bacteria for resource recovery: challenges and opportunities.Crossref | GoogleScholarGoogle Scholar |
[32] Hülsen, T et al. (2022) Creating value from purple phototrophic bacteria via single-cell protein production. Curr Opin Biotechnol 76, 102726.
| Creating value from purple phototrophic bacteria via single-cell protein production.Crossref | GoogleScholarGoogle Scholar |
[33] Razzak, SA et al. (2013) Integrated CO2 capture, wastewater treatment and biofuel production by microalgae culturing—a review. Renew Sustain Energy Rev 27, 622–653.
| Integrated CO2 capture, wastewater treatment and biofuel production by microalgae culturing—a review.Crossref | GoogleScholarGoogle Scholar |
[34] Rabaey, K and Rozendal, RA (2010) Microbial electrosynthesis — revisiting the electrical route for microbial production. Nat Rev Microbiol 8, 706–716.
| Microbial electrosynthesis — revisiting the electrical route for microbial production.Crossref | GoogleScholarGoogle Scholar |
[35] IPCC (2022) Climate Change 2022: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (Pörtner H-O et al., eds). Cambridge University Press, Cambridge, UK, and New York, NY, USA.
| Crossref |



