Bioprospecting for and the applications of halophilic acidophiles in bioleaching operations
Melissa K. Corbett A and Elizabeth L. J. Watkin B *A Curtin Medical School, Curtin University, Kent Street, GPO Box U1987, Perth, WA 6845, Australia.
B School of Science, Edith Cowan University, 270 Joondalup Drive, Joondalup, WA 6027, Australia.

Dr Melissa Corbett is a Senior Lecturer at the Curtin University Medical School. As a teaching and research academic, she has been working in the field of bioleaching for 15 years, focussing on the solubilisation of phosphates ores for the recovery of rare earth elements, as well as bioprospecting for salt tolerant microorganisms. Alongside conducting research and supervising Honours and PhD students, she lectures undergraduate students in the field of microbiology and genetics. She has broad experience in environmental, medical and molecular microbiology. |

Elizabeth Watkin is Associate Dean (Science and Mathematics) and Professor of Environmental Microbiology in the School of Science, at Edith Cowan University. She has over 20 years of experience in the field of microbial ecology of environmental systems and covers the fields of mining biotechnology, mineral resource recovery and microbial fouling of water (particularly within mining systems). Her research team investigates biotechnological processes for environmental and industrial applications and approaches to mitigate microbially caused problems such as biocorrosion, biofouling and bioclogging. Prior to 2005, her main area of research was symbiotic nitrogen fixing bacteria in both agricultural and native legume species. She maintains a minor research interest in this area. |
Microbiology Australia 44(1) 45-48 https://doi.org/10.1071/MA23011
Submitted: 23 January 2023 Accepted: 5 February 2023 Published: 22 February 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
The economic recovery of metals from sulfide ores has become a topic of increasing interest due to the escalating demand for critical minerals and the reducing grade of available ores. Bioleaching is the use of acidophilic iron and sulfur-oxidising microorganisms to facilitate the extraction of base metals from primary sulfide ores and tailings. One significant issue limiting the use of bioleaching is the availability of freshwater due to the sensitivity of these microbes to chloride. The use of saline tolerant acidophilic iron- and-sulfur oxidising microorganisms will go a long way to addressing this issue. There are three possible means of sourcing suitable microorganisms; adaptation, genetic engineering and bioprospecting, with bioprospecting showing the greatest possibilities. Bioprospecting in search of native organisms for bioleaching operations has led researchers to numerous locations around the world and the isolation of iron- and sulfur-oxidising acidophiles that are capable of tolerating high levels of salinity has been of particular interest in these investigations.
Keywords: acid saline lakes, Acidihalobacter, acidophiles, bioleaching, bioprospecting, genetic engineering, halophiles, saline drains.
As the global transition to, and the goal of governments for a zero emissions energy system are embraced worldwide, the demand for critical minerals such as cobalt, copper, nickel, lithium and rare earth elements is outstripping current availability.1 Compounding the lack of materials are issues of low-grade ores (uneconomical using current recovery processes), water consumption and downstream co-contamination of unwanted elements such as uranium during traditional extraction processes. Commercial success of mining operations often includes reclaiming and recycling mine wastes with some sites operating bioleaching heaps as an inexpensive alternative to the traditional pyro-metallurgical methods for the extraction of metals from low grade sulfide ores.
Even though bioleaching has been employed successfully for decades, lengthy leaching cycles and low recovery efficiencies have hindered large scale investment and adoption of the process. The demand for critical minerals is increasing and the resulting upwards trajectory of mineral commodity prices showing no signs of slowing.2 As a result, investigations into optimising bioleaching practices for the reclamation of elements (re-processing) from fresh or abandoned tailings is gathering interest, as is the application of this technology to lower grade primary ores. To optimise bioleaching processes, numerous avenues are being explored to not only increase mineral yield from these practices, but also to decrease operating, environmental and maintenance costs of the complex systems, tackling different parameters of the procedures involved. Prospective strategies for advancing bioleaching operations include deliberate adaptation of organisms to extreme conditions, genetic engineering and bioprospecting.
The microorganisms currently applied to bioleaching of sulfide ores are consistently a combination of acidophilic bacteria and archaea whose ability to prosper in low pH conditions enables the dissolution of iron and inorganic sulfur by way of metabolism and acid generation, thus liberating recoverable minerals. Under mesophilic conditions, bioleaching populations are dominated by genera such as Acidithiobacillus, Acidiphilium, Acidiferrobacter and Leptospirillum,3 whereas increasing temperatures (up to 60°C) alters the taxa to favour Sulfobacillus, Acidimicrobium and Ferroplasma. Communities comprising multiple species are more adept at undertaking a variety of tasks than a single species alone,4 but all can be adversely affected by changes in numerous parameters including water potential, ore porosity and surface area undesirably resulting in decreased retrieval efficiencies.
These bioleaching microorganisms were originally isolated from some of the most inhospitable regions on earth5 so adaptive culturing of both pure and mixed populations to extreme conditions (high compound metal concentrations, acidity, salinity) has seen some successes. Sub-culturing under situations of continuous or accumulative ore concentrations has been adopted by many researchers as a way to enhance bioleaching performance and increase element recovery.6 However, it can be a difficult and lengthy process maintaining discrete population numbers overtime in these adapted cultures. An alternative approach to the adaptation of microbial populations is to utilise a naturally occurring microbial consortium. A consortium of microbes can have superior benefits over cultures of pure isolates as a broader array of multifaceted functions including inter-species biofilm formation occur that have demonstrated a greater rate of mineral solubilisation7 than when pure cultures were applied.
Advances in proteomic and metabolomic analysis of microbial systems has enabled researchers to understand how microorganisms can tolerate extreme conditions. With this information, genetic manipulation and modification of existing bioleaching microorganisms to improve on leaching efficiencies has shown promise when applied to the recovery of rare earth elements.8 The incorporation of metabolic pathways involved in the degradation of organic compounds from heterotrophs such as Acidiphilum or Sulfobacillus into autotrophic iron and sulfur oxidisers (Acidithiobacillus) could reduce issues of organic toxicity that organics often bring to bioleaching operations.9 Designing and manufacturing synthetic microbial consortia for bioleaching applications is an emerging area of research, however, their application for in situ bioleaching (in particular heap leaching) remains to be seen because of strong environmental release laws. As microbial diversity, complete with wide metabolic potential, is influenced not only by the variable environment but also with the interactions between competing microorganisms, the genetic analysis of organisms often conducted on pure cultures does not reflect real world situations and should be taken into consideration if designing a synthetic population.
Bioleaching processes at high temperatures (>45°C) has seen success with the application of various thermophilic microbial species (Sulfolobus, Metallosphaera, Acidianus)10 for the recovery of copper, uranium and gold. Owing to the higher running costs of these operations (stirred tank reactors), this process is often restricted to high value minerals. This, nevertheless, has fuelled the search for more thermophilic organisms capable of solubilising iron–sulfide rich ores in locales such as hydrothermal vents11 (which is now an emerging deep-sea biotechnology industry), volcanic areas12 and hot springs.13
For bioleaching operations to be economically and environmentally sustainable, water consumption must be tightly controlled as it is essential for ore processing and recovery. Implementation of untreated groundwater sources for bioleaching applications runs the risk of tapping into sources with high levels of total dissolved solids, and the use of sea water requires understanding how chloride ions can affect mixed microbial communities.14 The mesophilic acidophiles listed earlier all demonstrate extreme sensitivity to chloride. Their cell membrane is permeable to the chloride ion, which, on entry to the cell, results in the negation of the positive membrane potential.15 Consequently, proton entry follows resulting in acidification of the cytoplasm, disturbing the proton motive force and eventually cell death. Microorganisms capable of surviving conditions of low pH, high salt and utilising Fe/S for energy are few in number as environments where these stresses co-exist are extraordinary unique. Therefore, bioprospecting for microorganisms that thrive in these conditions rapidly narrows the number of locations across the globe in which they may be found.
Our initial explorations took us to the acidic saline drains in the Yilgarn Craton of Western Australia in 2008 where we isolated a potential candidate known as F5.16 F5 effectively released base metals from pyrite, pentlandite and chalcopyrite under leaching conditions with up to 30 g L−1 of chloride ions present, and following genomic sequencing was named Acidihalobacter prosperus.17 This organism was then further re-classified as A. yilgarnensis due to genomic sequence comparisons with related species demonstrating a clear difference between it and an already existing A. prosperus.18 Acidihalobacter prosperus (originally named Thiobacillus prosperus) was isolated from a shallow geothermally heated seafloor on the Aeolian Islands, Vulcano, Italy.19 Subsequently a further two halotolerant acidophiles of this genus, A. prosperus V6 and A. ferrooxidans V8, were isolated from mixed shallow acidic marine pools, also on the Aeolian Islands. These isolates have subsequently been renamed A. prosperus, A. aeolianus and A. ferrooxydans,20,21 all of which are capable of growth at low pH (1.8–2.0) as well as tolerating high salt and oxidising pyritic compounds. The Acidihalobacter species may in the future be applied to bioleaching operations where concentrations of salt in the water render the usual consortium unviable.
Even though the organisms that currently make up the Acidihalobacter genus were isolated in locations greater than 13 000 km apart, similar environmental pressures have resulted in the conservation of genes essential for survival. Genome exploration of these organisms has revealed that the low pH and high salt tolerances evolved separately22 with the halophilic organism gaining genes for acid tolerance through horizontal gene transfer. Armed with this information, in vitro modelling of genetic alterations made to bioleaching organisms (Acidithiobacillus ferrooxidans) can allow us to predict the success and applicability of organisms for mineral recovery in high salt, low pH conditions.
As the Yilgarn Craton of Western Australia is dotted with numerous acidic saline lakes, it provides an ideal opportunity for further prospecting23 while utilising the information gained from molecular modelling to target specific environments with the greatest chance of hosting prokaryotes suitable for bioleaching in regions where saline water is of concern. Another hypersaline lake in Australia whose pH fluctuates with the seasons is Lake Tyrell (a shallow, salt-crusted depression) in Victoria, a location where numerous bioprospecting expeditions have been conducted24,25 in attempt to isolate organisms for biotechnological applications. A global search of other hypersaline lakes with low pH (<4) for iron–sulfur-oxidising halophiles to either adapt to acidic conditions or genetically modify to contain acid resistance mechanisms could allow for the construction of a consortia suitable for bioleaching in regions with salt contaminated waters (Fig. 1).
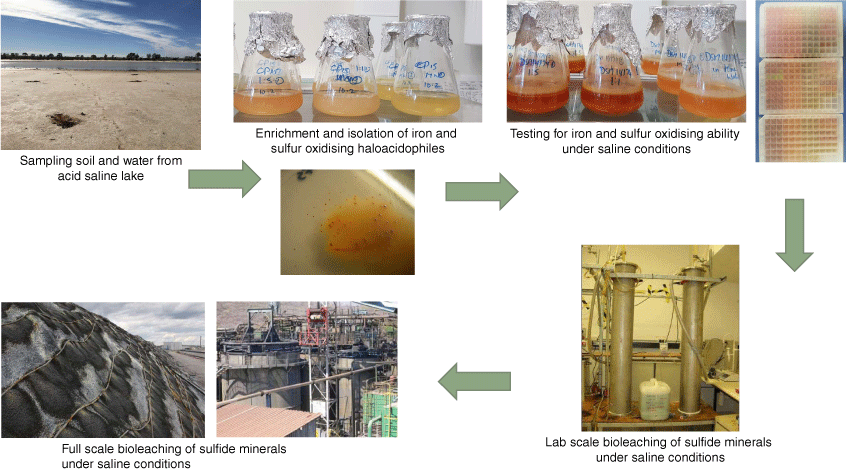
|
By understanding the microbial diversity, proteomic, lipidomic and genetic contributions, adaptations and pressures in the unique high salt, low pH environments, advances in the efficiency of bioleaching process could be accomplished. Although a range of approaches such as adaption and genetic modification could be applied to answering this problem, bioprospecting is the approach most likely to provide a successful solution.
Data availability
Data sharing is not applicable as no new data were generated or analysed during this study.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This research did not receive any specific funding.
References
[1] Hund K et al. (2022) Minerals for Climate Action: The Mineral Intensity of the Clean Energy Transition. International Bank for Reconstruction and Development and The World Bank, Washington, DC, USA.[2] Kim T-Y (2022) Scale of price increase for selected energy transition minerals and metals: IEA Org. https://www.iea.org/commentaries/critical-minerals-threaten-a-decades-long-trend-of-cost-declines-for-clean-energy-technologies
[3] Vera, M et al. (2022) Progress in bioleaching: fundamentals and mechanisms of microbial metal sulfide oxidation – part A. Appl Microbiol Biotechnol 106, 6933–6952.
| Progress in bioleaching: fundamentals and mechanisms of microbial metal sulfide oxidation – part A.Crossref | GoogleScholarGoogle Scholar |
[4] Brune, KD and Bayer, TS (2012) Engineering microbial consortia to enhance biomining and bioremediation. Front Microbiol 3, 203.
| Engineering microbial consortia to enhance biomining and bioremediation.Crossref | GoogleScholarGoogle Scholar |
[5] Chen, L-x et al. (2016) Microbial communities, processes and functions in acid mine drainage ecosystems. Curr Opin Biotechnol 38, 150–158.
| Microbial communities, processes and functions in acid mine drainage ecosystems.Crossref | GoogleScholarGoogle Scholar |
[6] Wang, Y et al. (2014) A moderately thermophilic mixed microbial culture for bioleaching of chalcopyrite concentrate at high pulp density. Appl Environ Microbiol 80, 741–750.
| A moderately thermophilic mixed microbial culture for bioleaching of chalcopyrite concentrate at high pulp density.Crossref | GoogleScholarGoogle Scholar |
[7] Corbett, MK et al. (2018) Syntrophic effect of indigenous and inoculated microorganisms in the leaching of rare earth elements from Western Australian monazite. Res Microbiol 169, 558–568.
| Syntrophic effect of indigenous and inoculated microorganisms in the leaching of rare earth elements from Western Australian monazite.Crossref | GoogleScholarGoogle Scholar |
[8] Schmitz, AM et al. (2021) Generation of a Gluconobacter oxydans knockout collection for improved extraction of rare earth elements. Nat Commun 12, 6693.
| Generation of a Gluconobacter oxydans knockout collection for improved extraction of rare earth elements.Crossref | GoogleScholarGoogle Scholar |
[9] Chen, J et al. (2022) Harnessing synthetic biology for sustainable biomining with Fe/S-oxidizing microbes. Front Bioeng Biotechnol 10, 920639.
| Harnessing synthetic biology for sustainable biomining with Fe/S-oxidizing microbes.Crossref | GoogleScholarGoogle Scholar |
[10] Watling, HR et al. (2016) Saline-water bioleaching of chalcopyrite with thermophilic, iron(II)- and sulfur-oxidizing microorganisms. Res Microbiol 167, 546–54.
| Saline-water bioleaching of chalcopyrite with thermophilic, iron(II)- and sulfur-oxidizing microorganisms.Crossref | GoogleScholarGoogle Scholar |
[11] Liu, W et al. (2020) Microbial imprints on sulfide minerals in submarine hydrothermal deposits of the East Pacific Rise. J Geophys Res Biogeosci 125, e2020JG005736.
| Microbial imprints on sulfide minerals in submarine hydrothermal deposits of the East Pacific Rise.Crossref | GoogleScholarGoogle Scholar |
[12] Hofmann, M et al. (2022) Metallosphaera javensis sp. nov., a novel species of thermoacidophilic archaea, isolated from a volcanic area. Int J Syst Evol Microbiol 72, 005536.
| Metallosphaera javensis sp. nov., a novel species of thermoacidophilic archaea, isolated from a volcanic area.Crossref | GoogleScholarGoogle Scholar |
[13] Kozubal, MA et al. (2012) Microbial iron cycling in acidic geothermal springs of yellowstone national park: integrating molecular surveys, geochemical processes, and isolation of novel fe-active microorganisms. Front Microbiol 3, 109.
| Microbial iron cycling in acidic geothermal springs of yellowstone national park: integrating molecular surveys, geochemical processes, and isolation of novel fe-active microorganisms.Crossref | GoogleScholarGoogle Scholar |
[14] Martínez-Bellange, P et al. (2022) Biomining of metals: new challenges for the next 15 years. Microb Biotechnol 15, 186–188.
| Biomining of metals: new challenges for the next 15 years.Crossref | GoogleScholarGoogle Scholar |
[15] Falagán, C and Johnson, DB (2018) The significance of pH in dictating the relative toxicities of chloride and copper to acidophilic bacteria. Res Microbiol 169, 552–557.
| The significance of pH in dictating the relative toxicities of chloride and copper to acidophilic bacteria.Crossref | GoogleScholarGoogle Scholar |
[16] Zammit, CM et al. (2009) The characterization of salt tolerance in biomining microorganisms and the search for novel salt tolerant strains. Adv Mater Res 71–73, 283–286.
| The characterization of salt tolerance in biomining microorganisms and the search for novel salt tolerant strains.Crossref | GoogleScholarGoogle Scholar |
[17] Khaleque, HN et al. (2017) Complete genome sequence of Acidihalobacter prosperus strain F5, an extremely acidophilic, iron- and sulfur-oxidizing halophile with potential industrial applicability in saline water bioleaching of chalcopyrite. J Biotechnol 262, 56–59.
| Complete genome sequence of Acidihalobacter prosperus strain F5, an extremely acidophilic, iron- and sulfur-oxidizing halophile with potential industrial applicability in saline water bioleaching of chalcopyrite.Crossref | GoogleScholarGoogle Scholar |
[18] Khaleque, HN et al. (2020) Genome-based classification of Acidihalobacter prosperus F5 (=DSM 105917=JCM 32255) as Acidihalobacter yilgarnensis sp. nov. Int J Syst Evol Microbiol 70, 6226–6234.
| Genome-based classification of Acidihalobacter prosperus F5 (=DSM 105917=JCM 32255) as Acidihalobacter yilgarnensis sp. nov.Crossref | GoogleScholarGoogle Scholar |
[19] Huber, H and Stetter, KO (1989) Thiobacillus prosperus sp. nov., represents a new group of halotolerant metal-mobilizing bacteria isolated from a marine geothermal field. Arch Microbiol 151, 479–485.
| Thiobacillus prosperus sp. nov., represents a new group of halotolerant metal-mobilizing bacteria isolated from a marine geothermal field.Crossref | GoogleScholarGoogle Scholar |
[20] Cárdenas, JP et al. (2015) Reclassification of ‘Thiobacillus prosperus’ Huber and Stetter 1989 as Acidihalobacter prosperus gen. nov., sp. nov., a member of the family Ectothiorhodospiraceae. Int J Syst Evol Microbiol 65, 3641–3644.
| Reclassification of ‘Thiobacillus prosperus’ Huber and Stetter 1989 as Acidihalobacter prosperus gen. nov., sp. nov., a member of the family Ectothiorhodospiraceae.Crossref | GoogleScholarGoogle Scholar |
[21] Khaleque, HN et al. (2019) Genome-based classification of two halotolerant extreme acidophiles, Acidihalobacter prosperus V6 (=DSM 14174 =JCM 32253) and ‘Acidihalobacter ferrooxidans’ V8 (=DSM 14175 =JCM 32254) as two new species, Acidihalobacter aeolianus sp. nov. and Acidihalobacter ferrooxydans sp. nov., respectively. Int J Syst Evol Microbiol 69, 1557–1565.
| Genome-based classification of two halotolerant extreme acidophiles, Acidihalobacter prosperus V6 (=DSM 14174 =JCM 32253) and ‘Acidihalobacter ferrooxidans’ V8 (=DSM 14175 =JCM 32254) as two new species, Acidihalobacter aeolianus sp. nov. and Acidihalobacter ferrooxydans sp. nov., respectively.Crossref | GoogleScholarGoogle Scholar |
[22] Boase, K et al. (2022) Prediction and inferred evolution of acid tolerance genes in the biotechnologically important Acidihalobacter genus. Front Microbiol 13, 848410.
| Prediction and inferred evolution of acid tolerance genes in the biotechnologically important Acidihalobacter genus.Crossref | GoogleScholarGoogle Scholar |
[23] Conner, AJ and Benison, KC (2013) Acidophilic halophilic microorganisms in fluid inclusions in halite from Lake Magic, Western Australia. Astrobiology 13, 850–860.
| Acidophilic halophilic microorganisms in fluid inclusions in halite from Lake Magic, Western Australia.Crossref | GoogleScholarGoogle Scholar |
[24] Narasingarao, P et al. (2012) De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6, 81–93.
| De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities.Crossref | GoogleScholarGoogle Scholar |
[25] Heidelberg, KB et al. (2013) Characterization of eukaryotic microbial diversity in hypersaline Lake Tyrrell, Australia. Front Microbiol 4, 115.
| Characterization of eukaryotic microbial diversity in hypersaline Lake Tyrrell, Australia.Crossref | GoogleScholarGoogle Scholar |


