The importance of micromonosporae in tropical, sub-tropical, terrestrial and aquatic environments
İpek Kurtböke A *A

Assoc. Prof. İpek Kurtböke has been working in the field of biodiscovery and has been an active member of the international actinomycete research community since 1982. She currently conducts research and teaches in the field of environmental and applied microbiology and biotechnology at the University of the Sunshine Coast, Queensland. She has also been an active member of the World Federation of Culture Collections (WFCC) and currently is the President of the Federation. She was also an Editorial Board member of Microbiology Australia for 20 years (2004–2024). |
Abstract
The genus Micromonospora (Ørskov 1923) is a Gram-positive, chemo-organotrophic, aerobic actinomycete, the second most predominant genus of the phylum Actinomycetota in natural environments after the genus Streptomyces, and has provided many clinically significant antibiotics, mostly aminoglycosides like gentamicin. This mini review will highlight the occurrence of this genus in diverse environments and its importance for natural product discovery and biotechnology.
Morphology and growth
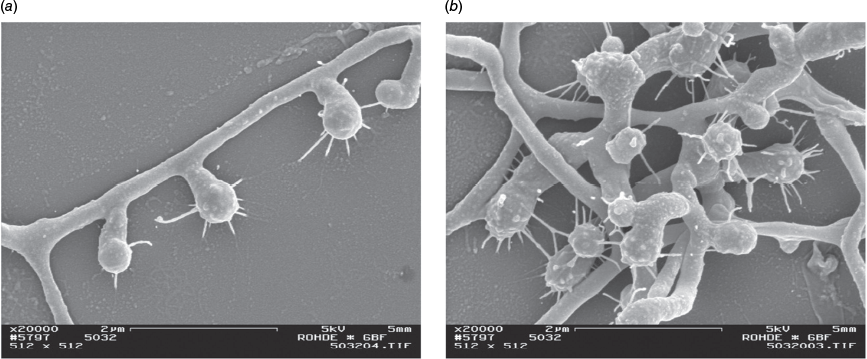
The genus Micromonospora belongs to the family Micromonosporaceae and the order Micromonosporales of the class Actinomycetes and the phylum Actinomycetota. It currently has 168 species, nine subspecies and one variety (https://lpsn.dsmz.de/). Species have extensive substrate mycelium (0.2–0.6 µm diameter), and non-motile spores are borne on short hyphae of the substrate mycelium1 (Fig. 1). Spores can be spherical to oval (0.7–1.5 µm), and in most species, blunt spiny projections are formed (Fig. 2a, b). Aerial mycelium is absent, but some cultures were reported to produce sterile short hyphae upon aging.1
Single spore clusters sitting on an extensively developed substrate mycelium (©Kurtböke personal collection).

Their spores are relatively resistant to physical and chemical treatments and unaffected by high heat and desiccation.2 On growth media, they form yellow, orange, red, brown, blue-green or purple colonies, but upon sporulation the colour can turn into dark brown or black (Fig. 3).3,4 Spores can remain viable in soil, lake and marine sediments for hundreds of years, and they can tolerate reduced oxygen tensions; as a result, they can remain viable in marine sediments.2,4
Ecology
Micromonospora are chemo-organotrophic and grow in aerobic and microaerophilic conditions. They can be sensitive to lower pH and optimum growth temperature ranges between 20°C and 40°C. They are mostly mesophilic, but they have been isolated from thermal hot springs in China5 and the Mongolian desert, where temperatures can reach thermophilic conditions.6 Kurapova et al. (2008)7 reported the presence of thermotolerant Micromonospora from the desert-steppe soil samples of Mongolia and the mountainous meadow soils of the Central Caucasus. They also found the representatives of the genus Micromonospora predominating on cyanobacterial biofilms on volcanic ash near hot springs of Kamchatka.7 Micromonospora were also detected in soils of the Cisolok geothermal area in West Java, Indonesia.8 Mehetre et al. (2019)9 reported the presence of micromonosporae in the Unkeshwar hot springs of India, where the temperature of the water of the hot springs ranges between 50°C and 60°C with pH values ranging between 6.9 and 7.2. A new species named M. halotolerans growing in the presence of 5% NaCl was found in the rhizosphere of a Pisum sativum plant from Spain.10 Zenova et al. (2011)11 reported the predominance of acidophilic species of the genus Micromonospora in acidic soils such as peat, soddy podzolic and taiga podzol.
Mangroves of subtropical and tropical environments
Mangrove environments have been reported to be a prolific source for micromonosporae, including the discovery of many new species of the genus. Examples include M. avicenniae sp. nov., isolated from a root of Avicennia marina, or M. rhizosphaerae sp. nov., isolated from mangrove Excocaria agallocha rhizosphere soil of a mangrove forest in the Hainan province of China.12,13 In particular, the Southeast Asia Pacific region micromonosporae were detected in mangrove environments in countries like China, India, Indonesia, Malaysia, Thailand, Vietnam and Australia.14–20
Bioactivity of micromonosporae
Antibiotics
Starting from the early 1960s many key antibiotics were discovered from this taxon, and Micromonospora species have been prolific producers of aminoglycoside antibiotics.21,22 Key antibiotics from this genus are listed in Table 1.
| Antibiotic class | ||||
|---|---|---|---|---|
| Antibiotic/year | Producer species | References | ||
| Aminoglycosides | Gentamicin (1963) | M. purpurea (NRRL 2953) and M. echinospora (NRRL 2985) and two subspecies, M. echinospora subsp. jerruginea (NRRL 2995) and subsp. pallida (NRRL 2996). | 23 | |
| Sisomycin (1970) | M. inyoensis (NRRL 3292). | 24, 25 | ||
| Verdamicin (1975) | M. grisea (NRRL 3800). | 26 | ||
| Fortimicin (1976) | M. olivoasterospora (ATCC 21819,31010,31009). | 27 | ||
| Neomycin (1973) | Micromonospora sp. (69–683) | 28 | ||
| Sagamicin (1974) | M. sagamiensis (ATCC 21803 and ATCC 21826) | 29 | ||
| Macrolides | Megalomicins (1969) | M. megalomicea (M. megalomicea var. megalomicea (NRRL 3274) and M megalomicea var. nigra (NRRL 3275) | 30 | |
| Rosaramicin (1972) | M. rosario NRRL 3178 | 31 | ||
| Juvenimicins (1976) | M. chalcea var. izumensis (ATCC 21561) | 32 | ||
| Erythromycin B (1976) | Micromonospora sp. (1225) | 33 | ||
| Ansamysins | Halomicin (1967) | M. halophytica (NRRL 2998) and M halophytica var. nigra (NRRL 3097) | 34 | |
| Rifamycin (1975) | M ellipsospora 71372 (NRRL 8021) | 35 | ||
| Everninomicins | Everninomicin complex (1965) | M. carbonacea (NRRL 2972) and M. carbonacea var. aurantiaca (NRRL 2997) | 36 | |
| Actinomycins | Actinomycin complex (1976) | M. floridensis (NRRL 8020) | 37 | |
| Miscellaneous antibiotics | Micromonospora species | 38 | ||
Note: Table information extracted from Wagman and Weinstein (1980).38
Micromonosporae still continue to provide novel antimicrobial compounds.39 He et al. (2001)40 reported Lomaiviticins A and B, which were potent antitumor antibiotics identified from M. lomaivitiensis isolated from the inner core of an ascidian. Back et al. (2021)41 again isolated a new Micromonospora strain (28ISP2-46T), recovered from the microbiome of a mid-Atlantic deep-sea sponge. Their whole-genome sequencing revealed that the bacterium could produce a diverse array of natural products, including kosinostatin and isoquinocycline B, which exhibited both antibiotic and antitumour properties. Genomics-driven approaches are now taking the discoveries to a new frontier; examples include the identification of a benzoxazole alkaloid-encoding biosynthetic gene cluster (mich BGC) by Cheng et al. (2023)42 in a marine-derived Micromonospora sp. (SCSIO 07g95). They subsequently, via the use of heterologous expression of this BGC in Streptomyces albus, discovered five new benzoxazole alkaloids, microechmycin A–E. Kokkini et al. (2022), using the combination of LC-UV-HRMS analyses, metabolomics analysis and molecular networking (GNPS) revealed that micromonosporae in their collection produced several related spirotetronates not disclosed before, including two new phocoenamicins, phocoenamicins D and E, belonging to this class of compounds.43,44
Bioactivity from mangrove environment-associated micromonosporae has also been reported. Examples include Ismet et al. (2004)45 who isolated and chemically characterised antifungal metabolites from Micromonospora sp. (M39) isolated from mangrove rhizosphere soil. Sarveswari et al. (2020)46 reported a mangrove-derived Micromonospora sp. (RMA46) as a potential source of anti-infectives against Vibrio cholerae. Wang et al. (2019)47 identified three new isoflavonoid glycosides from the mangrove-derived M. aurantiaca (110B). Pradhan and Das (2025)48 in India, Anggelina et al. (2021)49 in Indonesia, and Lee et al. (2014)50 in Malaysia detected bioactive micromonosporae in mangrove sediments. Antimicrobial activities were also detected from Micromonospora strains isolated from Thai soils51 as well as from the stains isolated from Ninh Thuan and Binh Thuan Seas of Vietnam.52
Enzymes
Simmons et al. (2014)53 analysed meta-transcriptomes from microbial communities and, using differential expression analysis, they identified enzymes, and they noted that overexpression of these protein families in the thermophilic community resulted from expression of a small number of genes not currently represented in any protein database. Interestingly, genes in overexpressed protein families were predominantly expressed by a single actinomycete genus, which was Micromonospora. They concluded that Micromonospora dominates expression of lignocellulolytic enzymes in a thermophilic community, thus making the members of this genus a promising source of lignocellulolytic enzymes for biotechnological applications. Kumas et al. (2023)54 also agreed with Simmons et al. (2014)53 and their analysis of protein family representation in each metagenome they studied indicated that cellobiohydrolases were significantly overrepresented, and the members of the genus Micromonospora primarily housed these genes.
Amylase, lipase and urease activities were found from micromonosporae associated with mangroves in the Setiu Wetland in Terengganu in Malaysia.55 Cellulase enzymes were detected from micromonospora species and used to control plant pathogenic fungi by El-Tarabily et al. (1996).56 Again, El-Tarabily et al. (2000)57 reported production of chitinase and β-1,3-glucanase from M. carbonacea that caused extensive plasmolysis and cell wall lysis of Sclerotinia minor hyphae in vitro and in glass house tests. Termite gut-associated actinomycetes, including micromonosporae have also been reported as sources of bioactive compounds and enzymes.58
Tiong et al. (2023)59 noted that one of the exceptionally heat-resistant enzymes capable of degrading PET is the leaf-branch compost cutinase (LCC) and using molecular advances (e.g. genomic mining and in silico analysis of distant cutinase homologues), potential PET-degrading hydrolases can be obtained from actinomycetes, including from the members of Micromonospora.
Other biotechnologically important natural products
Abdelmohsen et al. (2012)60 reported antioxidant and anti-protease activities of diazepinomicin from the sponge-associated Micromonospora strain (RV115). Vitamin B12 was also reported to be produced by the members of this genus (Wagman et al. 1969).61
Conclusions
The second most predominant genus of the class Actinomycetes after the genus Streptomyces, Micromonospora will stay as one of the most important genera for biodiscovery and biotechnology,21 including the production of biofuels. Demain62 stated that one Micromonospora strain can produce 48 aminocyclitol antibiotics. The antibiotics vary in size from small molecules like cycloserine (102 Da) and bacilysin (270 Da) to polypeptides such as nisin, which contains 34 amino acid residues. With the advances in molecular techniques such as genome-based classification of micromonosporae with a focus on their biotechnological and ecological potential Carro et al. (2018)63 reported previously unrealised potential to synthesise novel specialised metabolites. They also noted that micromonosporae can adapt to key environmental variables via the distribution of key stress-related genes. Moreover, genes associated with plant interactions can open new avenues for the potential use of micromonosporae in agriculture and biotechnology.63 As stated by further investigations, such as the use of a multi-omics framework combining genome mining and metabolomics to explore the biosynthetic potential of wild-type strains of micromonosporae from extreme habitats, will reveal unknown potentials of this genus.64,65
Acknowledgements
The author thanks Professor Manfred Rohde, GBF, Germany, for his invaluable assistance with the SEM imagery (Figs 1, 2a, b).
References
5 Liu L, Salam N, Jiao JY, Jiang HC, Zhou EM, Yin YR, Ming H, Li WJ (2016) Diversity of culturable thermophilic actinobacteria in hot springs in Tengchong, China and studies of their biosynthetic gene profiles. Microb Ecol 72(1), 150-162.
| Crossref | Google Scholar | PubMed |
6 Kurapova AI, Zenova GM, Sudnitsyn II, Kizilova AK, Manucharova NA, Norovsuren Zh, Zvyagintsev DG (2012) Thermotolerant and thermophilic actinomycetes from soils of Mongolia desert steppe zone. Microbiology 81(1), 98-108.
| Crossref | Google Scholar |
7 Kurapova AI, Zenova GM, Orleanskii VK, Manucharov AS, Norovsuren Zh (2008) Mesophilic and thermotolerant actinomycetes in strongly heated soils. Mosc Univ Soil Sci Bull 63(3), 142-147.
| Crossref | Google Scholar |
8 Ningsih F, Sari DC, Rachmania MK, Yabe S, Yokota A, Oetari A, Sjamsuridzal W (2020) Isolation and 16S rRNA gene sequences analysis of thermophilic Actinobacteria isolated from soil in Cisolok geothermal area, West Java, Indonesia. IOP Conf Ser: Earth Enrivon Sci 457(1), 012015.
| Google Scholar |
9 Mehetre GT, Vinodh JS, Burkul BB, Desai D, Santhakumari B, Dharne MS, Dastager SG (2019) Bioactivities and molecular networking-based elucidation of metabolites of potent actinobacterial strains isolated from the Unkeshwar geothermal springs in India. RSC Adv 9(17), 9850-9859.
| Crossref | Google Scholar | PubMed |
10 Carro L, Pukall R, Spröer C, Kroppenstedt RM, Trujillo ME (2013) Micromonospora halotolerans sp. nov., isolated from the rhizosphere of a Pisum sativum plant. Antonie Van Leeuwenhoek 103(6), 1245-1254.
| Crossref | Google Scholar | PubMed |
11 Zenova GM, Manucharova NA, Zvyagintsev DG (2011) Extremophilic and extremotolerant actinomycetes in different soil types. Eurasian Soil Sci 44(4), 417-436.
| Crossref | Google Scholar |
12 Li L, Mao YJ, Xie QY, Deng Z, Hong K (2013) Micromonospora avicenniae sp. nov., isolated from a root of Avicennia marina. Antonie Van Leeuwenhoek 103(5), 1089-1096.
| Crossref | Google Scholar | PubMed |
13 Wang C, Xu XX, Qu Z, Wang HL, Lin HP, Xie QY, Ruan JS, Hong K (2011) Micromonospora rhizosphaerae sp. nov., isolated from mangrove rhizosphere soil. Int J Syst Evol Microbiol 61(2), 320-324.
| Crossref | Google Scholar | PubMed |
14 Xie QY, Ren J, Li L, Li Y, Deng ZX, Hong K (2016) Micromonospora mangrovi sp. nov., isolated from mangrove soil. Antonie van Leeuwenhoek 109(4), 483-491.
| Crossref | Google Scholar | PubMed |
15 Rajkumar J, Swarnakumar NS, Sivakumar K, Thangaradjou T, Kannan L (2012) Actinobacterial diversity of mangrove environment of the Bhitherkanika mangroves, east coast of Orissa, India. Int J Sci Res Publ 2(2), 1-6.
| Google Scholar |
16 Hidayatullah AR, Sugihartati R, Handijatno D, Chusniati S, Maslachah L, Sarudji S (2020) Isolation of Actinomycetes from mangrove sediments at Ujung Pangkah, Gresik, Indonesia. Ecol Environ Conserv 26, 231-237.
| Google Scholar |
17 Smet A, Parameswari S, Vikineswary S (2002) Diversity of Micromonospora in Malaysian mangrove rhizosphere soil. Malays J Sci 21(1&2), 51-59.
| Google Scholar |
18 Phongsopitanun W, Kudo T, Ohkuma M, Pittayakhajonwut P, Suwanborirux K, Tanasupawat S (2016) Micromonospora sediminis sp. nov., isolated from mangrove sediment. Int J Syst Evol Microbiol 66(8), 3235-3240.
| Crossref | Google Scholar | PubMed |
19 Van Hop D, Sakiyama Y, Binh CT, Otoguro M, Hang DT, Miyadoh S, Luong DT, Ando K (2011) Taxonomic and ecological studies of actinomycetes from Vietnam: isolation and genus-level diversity. J Antibiot 64(9), 599-606.
| Crossref | Google Scholar | PubMed |
20 Eccleston GP, Brooks PR, Kurtböke DI (2008) The occurrence of bioactive micromonosporae in aquatic habitats of the Sunshine Coast in Australia. Mar Drugs 6(2), 243-261.
| Crossref | Google Scholar | PubMed |
21 Hirsch AM, Valdés M (2010) Micromonospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol Biochem 42(4), 536-542.
| Crossref | Google Scholar |
22 Bérdy J (2005) Bioactive microbial metabolites. J Antibiot (Tokyo) 58, 1-26.
| Crossref | Google Scholar | PubMed |
23 Weinstein MJ, Luedemann GM, Oden EM, Wagman GH (1963) Gentamicin, a new broad spectrum antibiotic complex. Antimicrob Agents Chemother (Bethesda) 161, 1-7.
| Google Scholar | PubMed |
24 Wagman GH, Testa RT, Marquez JA (1970) Antibiotic 6640. II Fermentation, isolation, and properties. J Antibiot (Tokyo) 23(11), 555-558.
| Crossref | Google Scholar | PubMed |
25 Weinstein MJ, Marquez JA, Testa RT, Wagman GH, Oden EM, Waitz JA (1970) Antibiotic 6640, a new Micromonospora-produced aminoglycoside antibiotic. J Antibiot (Tokyo) 23, 551-554.
| Crossref | Google Scholar | PubMed |
26 Weinstein MJ, Wagman GH, Marquez JA, Testa RT, Waitz JA (1975) Verdamicin, a new broad spectrum aminoglycoside antibiotic. Antimicrob Agents Chemother 7(3), 246-249.
| Crossref | Google Scholar | PubMed |
27 Nara T, Yamamoto M, Kawamoto I, Takayama K, Okachi R, Takasawa S, Sato T, Sato S (1977) Fortimicins A and B, new aminoglycoside antibiotics I. Producing organism, fermentation and biological properties of fortimicins. J Antibiot (Tokyo) 30(7), 533-540.
| Crossref | Google Scholar | PubMed |
28 Wagman GH, Marquez JA, Watkins PD, Bailey JV, Gentile F, Weinstein MJ (1973) Neomycin production by Micromonospora species 69-683. J Antibiot (Tokyo) 26(12), 732-736.
| Crossref | Google Scholar | PubMed |
29 Okachi R, Kawamoto I, Takasawa S, Yamamoto M, Sato S (1974) A new antibiotic XK-62-2 (Sagamicin) I. Isolation, physicochemical and antibacterial properties. J Antibiot (Tokyo) 27(10), 793-800.
| Crossref | Google Scholar | PubMed |
30 Weinstein MJ, Wagman GH, Marquez JA, Testa RT, Oden E, Waitz JA (1969) Megalomicin, a new macrolide antibiotic complex produced by Micromonospora. J Antibiot (Tokyo) 22(6), 253-258.
| Crossref | Google Scholar | PubMed |
31 Wagman GH, Waitz JA, Marquez J, Murawaski A, Oden EM, Testa RT, Weinstein MJ (1972) A new Micromonospora-produced macrolide antibiotic, rosamicin. J Antibiot (Tokyo) 25(11), 641-646.
| Crossref | Google Scholar | PubMed |
32 Hatano K, Higashide E, Shibata M (1976) Studies on Juvenimicin, a new antibiotic. I taxonomy, fermentation and antimicrobial properties. J Antibiot (Tokyo) 29(11), 1163-1170.
| Crossref | Google Scholar | PubMed |
34 Weinstein MJ, Luedemann GM, Oden EM, Wagman GH (1967) Halomicin, a new Micromonospora-produced antibiotic. Antimicrob Agents Chemother (Bethesda) 7, 435-441.
| Crossref | Google Scholar | PubMed |
36 Weinstein MJ, Luedemann GM, Oden EM, Wagman GH (1964) Everninomicin, a new antibiotic complex from Micromonospora carbonacea. Antimicrob Agents Chemother (Bethesda) 10, 24-32.
| Google Scholar | PubMed |
37 Wagman GH, Marquez JA, Watkins PD, Gentile F, Murawski A, Patel M, Weinstein MJ (1976) A new actinomycin complex produced by a Micromonospora species: fermentation, isolation, and characterization. Antimicrob Agents Chemother (Bethesda) 9(3), 465-469.
| Crossref | Google Scholar | PubMed |
38 Wagman GH, Weinstein MJ (1980) Antibiotics from Micromonospora. Ann Rev Microbiol 34, 537-557.
| Crossref | Google Scholar | PubMed |
39 Boumehira AZ, El-Enshasy HA, Hacène H, Elsayed EA, Aziz R, Park EY (2016) Recent progress on the development of antibiotics from the genus Micromonospora. Biotechnol Bioprocess Eng 21(2), 199-223.
| Crossref | Google Scholar |
40 He H, Ding WD, Bernan VS, Richardson AD, Ireland CM, Greenstein M, Ellestad GA, Carter GT (2001) Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaivitiensis. J Am Chem Soc 123(22), 5362-5363.
| Crossref | Google Scholar | PubMed |
41 Back CR, Stennett HL, Williams SE, Wang L, Ojeda Gomez J, Abdulle OM, Duffy T, Neal C, Mantell J, Jepson MA, Hendry KR, Powell D, Stach J, Essex-Lopresti AE, Willis CL, Curnow P, Race PR (2021) A new Micromonospora strain with antibiotic activity isolated from the microbiome of a mid-Atlantic deep-sea sponge. Mar Drugs 19(2), 105.
| Crossref | Google Scholar | PubMed |
42 Cheng Z, Zhang Q, Peng J, Zhao X, Ma L, Zhang C, Zhu Y (2023) Genomics-driven discovery of benzoxazole alkaloids from the marine-derived micromonospora sp. SCSIO 07395. Molecules 28(2), 821.
| Crossref | Google Scholar | PubMed |
43 Kokkini M, González Heredia C, Oves-Costales D, de la Cruz M, Sánchez P, Martín J, Vicente F, Genilloud O, Reyes F (2022) Exploring Micromonospora as phocoenamicins producers. Mar Drugs 20(12), 769.
| Crossref | Google Scholar | PubMed |
44 Kokkini M, Oves-Costales D, Sánchez P, Melguizo Á, Mackenzie TA, Pérez-Bonilla M, Martín J, Giusti A, de Witte P, Vicente F, Genilloud O, Reyes F (2023) New phocoenamicin and maklamicin analogues from cultures of three marine-derived Micromonospora strains. Mar Drugs 21(8), 443.
| Crossref | Google Scholar | PubMed |
45 Ismet A, Vikineswary S, Paramaswari S, Wong WH, Ward A, Seki T, Fiedler HP, Goodfellow M (2004) Production and chemical characterization of antifungal metabolites from Micromonospora sp. M39 isolated from mangrove rhizosphere soil. World J Microbiol Biotechnol 20(5), 523-528.
| Crossref | Google Scholar |
46 Sarveswari HB, Kalimuthu S, Shanmugam K, Neelakantan P, Solomon AP (2020) Exploration of anti-infectives from mangrove-derived Micromonospora sp. RMA46 to combat Vibrio cholerae pathogenesis. Front Microbiol 11, 1393.
| Crossref | Google Scholar | PubMed |
47 Wang RJ, Zhang SY, Ye YH, Yu Z, Qi H, Zhang H, Xue ZL, Wang JD, Wu M (2019) Three new isoflavonoid glycosides from the mangrove-derived actinomycete Micromonospora aurantiaca 110B. Mar Drugs 17(5), 294.
| Crossref | Google Scholar | PubMed |
48 Pradhan D, Das MT (2025) Antimicrobial potential and extracellular metabolite profiling of bichitrapur mangrove sediment derived Micromonospora sp. BSS-D-04 and Streptomyces sp. BSS-D-05 using LC–MS/MS and GC–MS analysis. Braz J Microbiol 2025, 1-17.
| Crossref | Google Scholar | PubMed |
49 Anggelina AC, Pringgenies D, Setyati WA (2021) Presence of biosynthetic gene clusters (nrps/pks) in actinomycetes of mangrove sediment in Semarang and Karimunjawa, Indonesia. Environ Nat Resour J 19(5), 391-401.
| Crossref | Google Scholar |
50 Lee LH, Zainal N, Azman AS, Eng SK, Goh BH, Yin WF, Ab Mutalib NS, Chan KG (2014) Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci World J 2014(1), 698178.
| Crossref | Google Scholar | PubMed |
51 Songsumanus A (2013) Characterization and screening of antimicrobial activity of Micromonospora strains from Thai soils. Malays J Microbiol 9(3), 260-269.
| Crossref | Google Scholar |
52 Vu TTH (2022) Screening of actinomycetes from Ninh Thuan and Binh Thuan Seas for antimicrobial producers. Vietnam J Mar Sci Technol 22(4), 423-432.
| Crossref | Google Scholar |
53 Simmons CW, Reddy AP, D'haeseleer P, Khudyakov J, Billis K, Pati A, Simmons BA, Singer SW, Thelen MP, VanderGheynst JS (2014) Metatranscriptomic analysis of lignocellulolytic microbial communities involved in high-solids decomposition of rice straw. Biotechnol Biofuels 7(1), 495.
| Crossref | Google Scholar | PubMed |
54 Kumas A, Ertekin SG, Gurbanov R, Simsek YE, Kocak FO (2023) Effect of Micromonospora sp. KSC08 on nitrogen conservation throughout composting. Biomass Convers Biorefin 13(3), 2375-2390.
| Crossref | Google Scholar |
55 Nor Hasan HN, Abdullah MDD, Saidin JB (2024) Antimicrobial and enzymatic activities of mangrove-associated actinomycetes. Malays Appl Biol 53(3), 219-228.
| Crossref | Google Scholar |
56 El-Tarabily KA, Sykes ML, Kurtböke ID, Hardy GESJ, Barbosa AM, Dekker RFH (1996) Synergistic effects of a cellulase-producing Micromonospora carbonaceae and an antibiotic-producing Streptomyces violascens on the suppression of Phytophthora cinnamomi root rot of Banksia grandis. Canad J Bot 74, 618-624.
| Crossref | Google Scholar |
57 El-Tarabily KA, Soliman MH, Nassar AH, Al-Hassani HA, Sivasithamparam K, McKenna F, Hardy GESJ (2000) Biological control of Sclerotinia minor using a chitinolytic bacterium and actinomycetes. Plant Pathol 49(5), 573-583.
| Crossref | Google Scholar |
58 Kurtböke Dİ, French, JRJ, Hayes, RA, Quinn, RJ. Eco-taxonomic insights into actinomycete symbionts of termites for discovery of novel bioactive compounds. In: Mukherjee J, editor. Biotechnological Applications of Biodiversity. Advances in Biochemical Engineering/Biotechnology. Vol. 147. Berlin Heidelberg: Springer-Verlag; 2014. pp. 111–135. 10.1007/10_2014_270
59 Tiong E, Koo YS, Bi J, Koduru L, Koh W, Lim YH, Wong FT (2023) Expression and engineering of PET-degrading enzymes from Microbispora, Nonomuraea, and Micromonospora. Appl Environ Microbiol 89(11), e00632-23.
| Crossref | Google Scholar | PubMed |
60 Abdelmohsen UR, Szesny M, Othman EM, Schirmeister T, Grond S, Stopper H, Hentschel U (2012) Antioxidant and anti-protease activities of diazepinomicin from the sponge-associated Micromonospora strain RV115. Mar Drugs 10(10), 2208-2221.
| Crossref | Google Scholar | PubMed |
61 Wagman GH, Gannon RD, Weinstein MJ (1969) Production of vitamin B12 by Micromonospora. Appl Microbiol 17(4), 648-649.
| Crossref | Google Scholar | PubMed |
62 Demain AL (2000) Small bugs, big business: the economic power of the microbe. Biotechnol Adv 18(6), 499-514.
| Crossref | Google Scholar | PubMed |
63 Carro L, Nouioui I, Sangal V, Meier-Kolthoff JP, Trujillo ME, Montero-Calasanz M, Sahin N, Smith DL, Kim KE, Peluso P, Deshpande S, Woyke T, Shapiro N, Kyrpides NC, Klenk HP, Göker M, Goodfellow M (2018) Genome-based classification of micromonosporae with a focus on their biotechnological and ecological potential. Sci Rep 8(1), 525.
| Crossref | Google Scholar | PubMed |
64 Han J-R, Li S, Lian W-H, Xu L, Duan L, Li J-L, Feng C-Y, Shi G-Y, Liu W-L, Wei Q-C, Li W-J, Dong L (2025) Multi‐omics analyses uncovering the biosynthetic potential of novel Micromonospora species isolated from desert and marine habitats. J Syst Evol
| Crossref | Google Scholar |
65 Yan S, Zeng M, Wang H, Zhang H (2022) Micromonospora: a prolific source of bioactive secondary metabolites with therapeutic potential. J Med Chem 65(13), 8735-8771.
| Crossref | Google Scholar | PubMed |
 Assoc. Prof. İpek Kurtböke has been working in the field of biodiscovery and has been an active member of the international actinomycete research community since 1982. She currently conducts research and teaches in the field of environmental and applied microbiology and biotechnology at the University of the Sunshine Coast, Queensland. She has also been an active member of the World Federation of Culture Collections (WFCC) and currently is the President of the Federation. She was also an Editorial Board member of Microbiology Australia for 20 years (2004–2024). |