Whole-genome sequencing as an improved means of investigating Neisseria gonorrhoeae treatment failures
Cameron Buckley A , Scott A. Beatson B C D , Athena Limnios E , Monica M. Lahra E F , David M. Whiley A B G and Brian M. Forde B C D HA Faculty of Medicine, UQ Centre for Clinical Research, The University of Queensland, Herston, Qld 4029, Australia.
B Australian Infectious Diseases Research Centre, The University of Queensland, Brisbane, Qld 4072, Australia.
C Australian Centre for Ecogenomics, The University of Queensland, Brisbane, Qld 4072, Australia.
D School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Qld 4072, Australia.
E WHO Collaborating Centre for STI and AMR, Microbiology Department, New South Wales Health Pathology, Prince of Wales Hospital, Sydney, NSW 2031, Australia.
F School of Medical Sciences, UNSW, Sydney, NSW 2052, Australia.
G Pathology Queensland, Microbiology Department, Herston, Qld 4029, Australia.
H Corresponding author: Email: b.forde@uq.edu.au
Sexual Health 16(5) 500-507 https://doi.org/10.1071/SH19012
Submitted: 24 January 2019 Accepted: 22 August 2019 Published: 4 September 2019
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
Background: Although rare, Neisseria gonorrhoeae treatment failures associated with ceftriaxone have been reported. The World Health Organization (WHO) recommends standardised protocols to verify these cases. Two cases from Australia were previously investigated using N. gonorrhoeae multiantigen sequence typing (NG-MAST), which has been used extensively to assess treatment failures. Case 1 pharyngeal isolates were indistinguishable, whereas Case 2 pharyngeal isolates were distinguished based on an 18-bp deletion in the major outer membrane porin encoded by the porB gene, questioning the reliability of NG-MAST results. Here we used whole-genome sequencing (WGS) to reinvestigate Cases 1 and 2, with a view to examining WGS to assess treatment failures. Methods: Pre- and post-treatment isolates for each case underwent Illumina sequencing, and the two post-treatment isolates underwent additional long-read sequencing using Pacific Biosciences. Sequence data were interrogated to identify differences at single nucleotide resolution. Results: WGS identified variation in the pilin subunit encoded by the pilE locus for both cases and the specific 18-bp porB deletion in Case 2 was confirmed, but otherwise the isolates in each case were indistinguishable. Conclusions: The WHO recommends standardised protocols for verifying N. gonorrhoeae treatment failures. Case 2 highlights the enhanced resolution of WGS over NG-MAST and emphasises the immediate effect that WGS can have in a direct clinical application for N. gonorrhoeae. Assessing the whole genome compared with two highly variable regions also provides a more confident predictor for determining treatment failure. Furthermore, WGS facilitates rapid comparisons of these cases in the future.
Additional keywords: antimicrobial resistance, public health, surveillance.
Introduction
Antimicrobial-resistant (AMR) Neisseria gonorrhoeae has become a major global public health concern. Ceftriaxone is considered the last remaining empirical monotherapy for gonorrhoea, but isolates exhibiting decreased susceptibility to ceftriaxone are widespread1 and there are recent reports of transmissible ceftriaxone-resistant strains from several countries.2–4 Although relatively rare, there have also been a handful of reports regarding treatment failure using ceftriaxone monotherapy.5–10 A dual therapy approach, combining azithromycin with ceftriaxone, as recommended by the World Health Organization (WHO), has recently been adopted in numerous countries as a means of delaying the emergence and spread of resistant gonorrhoea.11 Although there is some suggestion that dual therapy has stabilised or even decreased cephalosporin resistance,12 there have been concomitant widespread increases in azithromycin-resistant N. gonorrhoeae, as well as reports of strains now exhibiting resistance to both ceftriaxone and azithromycin.2,3 There have also been at least two confirmed treatment failures using the dual therapy approach.3,13
Among the strategies proposed in the WHO global action plan to minimise the impact of AMR gonorrhoea is the need to develop protocols to standardise and verify gonorrhoea treatment failures.14 At the laboratory level, this typically involves using phenotypic or genotypic tools including multilocus or N. gonorrhoeae multi-antigen sequence typing (MLST and NG-MAST respectively) to evaluate pre- and post-treatment isolates that are taken approximately 1 week apart. Isolates that are indistinguishable based on these traditional methods are considered treatment failures, whereas distinguishable isolates are considered consistent with reinfection. The major limitation of traditional gonococcal typing approaches is that they can have limited discriminatory power and may fail to distinguish different strains. Conversely, many target highly variable regions, like the gonococcal outer membrane protein which is encoded by the porB gene that may potentially mutate during therapy.
The NG-MAST method is a widely used gonococcal genotyping tool that involves DNA sequencing of the gonococcal transferrin-binding protein (which is encoded by tbpB) and porB gene, corresponding with 390 and 490 bp fragments, respectively.15 The method has been used extensively to examine treatment failures.16 In 2011, when ceftriaxone monotherapy was the recommended treatment for uncomplicated gonorrhoea, NG-MAST was used to investigate two suspected treatment failures in Australia, both involving pharyngeal infection.8 Upon having their initial samples taken, the individuals were asked to abstain from any further sexual intercourse and return for a ‘test of cure’ approximately 1 week later to determine whether the infection had resolved or not. Although isolates from Case 1 were indistinguishable by NG-MAST and were consistent with clinical information indicating treatment failure, the isolates from Case 2 were distinguished by NG-MAST based on an 18-bp deletion in the porB sequence of the post-treatment isolate and a difference in the ceftriaxone minimum inhibitory concentration (MIC; albeit within one doubling dilution). The patient from Case 2 denied sexual contact in the follow-up period, raising questions over reliability of the NG-MAST results. Ultimately, we categorised Case 2 as a suspected treatment failure.
The issues associated with Case 2 highlight the potential limitations of established laboratory methods for investigating gonococcal treatment failures; notably such issues have been observed in earlier suspected treatment failure cases.7 As identified by the WHO, addressing these issues is a priority, particularly in an environment where ceftriaxone-resistant strains are now emerging and rapid identification of treatment failures would be needed to facilitate timely public health responses. Whole-genome sequencing (WGS) is now being widely used to enhance N. gonorrhoeae epidemiological investigations, including the ability to determine direct or indirect transmission links between patients17 and assessing minimal nucleotide substitutions between multiple isolates from the same patient.17,18 A recent study by Harris et al.19 comprehensively showed the potential limitations of NG-MAST for examining N. gonorrhoeae molecular epidemiology, including the inability of NG-MAST to accurately associate isolates with clinically relevant phylogenetic clades attained via WGS analyses. By providing single-nucleotide resolution, WGS enables better discrimination between strains considered to be linked via NG-MAST.20 In this study, we used Cases 1 and 2 (described above) as examples to examine the utility of WGS to assess treatment failure. Furthermore, we sought to address whether short-read data provided sufficient resolution to aid in interpretation of these cases.
Methods
Isolate collection
This investigation assessed two pharyngeal isolates from each individual. The first isolate was collected before the administration of antibiotics (pre-treatment isolate) and the second after antibiotic therapy (post-treatment isolate). The pre-treatment isolates were collected upon patients’ initial visit in March and July of 2011 (Case 1 and 2 respectively). The post-treatment isolates were acquired after patients’ return visits 1 week later, at which time the pharynx was still N. gonorrhoeae positive in both cases.
Sequencing
All four pharyngeal isolates underwent short-read Illumina (San Diego, CA, USA) sequencing, and the two post-treatment isolates underwent additional long-read sequencing using Pacific Biosciences (PacBio; Menlo Park, CA, USA) to generate high-quality genomes for each case. Only one isolate from each case was selected for long-read sequencing because a closely related reference genome is sufficient for identifying variants among isolates in cases of treatment failure. It should be noted that the Illumina sequencing for the pre- and post-treatment isolates was conducted at different times during the course of this study, and that the isolates were subsequently sequenced at different institutions.
Briefly, for post-treatment isolates, original stock cultures were recovered from –70°C storage and plated onto LB agar, which was then incubated at 37°C with 5% CO2 in air for 24 h. A single colony was selected and subcultured onto LB agar and incubated under the conditions described above. This culture was used for both PacBio and Illumina sequencing. Genomic (g) DNA was extracted using the Ultraclean Microbial DNA Isolate Kit (GeneWorks, Adelaide, SA, Australia) according to the manufacturer’s instructions. These post-treatment DNA extracts underwent sequencing using a PacBio RSII sequencer with P6-C4 chemistry by an external sequencing service provider (Doherty Institute, The University of Melbourne, Vic., Australia). For Illumina sequencing of the post-treatment isolates, libraries were prepared using the Nextera XT DNA kit (Illumina) and sequenced on the NextSeq 500 using the High Output v2 kit (Illumina) at the Australian Centre for Ecogenomics, University of Queensland.
The pre-treatment isolates were cultured as described above, and DNA was extracted using the DSP DNA Mini Kit on the QIAsymphony SP (Qiagen, Valencia, CA, USA). The pre-treatment isolates were sequenced at Queensland Health Forensic and Scientific Services (Coopers Plains, Qld, Australia), using the same Illumina method as above, with the exception that the Mid Output v2 kit (Illumina) was used.
Assembly, annotation and variant detection
The PacBio raw sequence data were de novo assembled using the hierarchical genome assembly process (HGAP 3.0,21 using SMRT Analysis v2.3.0 by PacBio; https://www.pacb.com/products-and-services/analytical-software/smrt-analysis/; accessed 25 August 2019) and polished using Quiver (https://github.com/PacificBiosciences/GenomicConsensus; accessed 25 August 2019),21 both using default settings. The assembled chromosome for each isolate was circularised by screening the respective 5′ and 3′ ends to identify overlapping sequences using Contiguity (https://mjsull.github.io/Contiguity/; accessed 25 August 2019), which were then manually trimmed. The circularised genomes underwent additional reiterative polishing with Pilon v1.22 (https://github.com/broadinstitute/pilon; accessed 25 August 2019)22 by using the Illumina reads to resolve single nucleotide insertions and deletions. Genome assemblies were further corrected by manual inspection of the read pileups. Complete chromosomes were then annotated using Prokka v1.12 (https://github.com/tseemann/prokka; accessed 25 August 2019).23
The Illumina raw sequence reads were assessed for quality using FastQC v0.11.4 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/; accessed 26 August 2019) and were subsampled to 1 100 000 reads if necessary using seqtk (https://github.com/lh3/seqtk; accessed 26 August 2019) to achieve comparable genome coverage for all isolates. A minimum read length of 100 bases was kept, and bases with a quality score <10 were removed using Trimmomatic v0.36 (https://github.com/timflutre/trimmomatic; accessed 26 August 2019).24 De novo assembly was achieved using SPAdes v3.10.1 (https://github.com/ablab/spades; accessed 26 August 2019)25 with the ‘–careful’ flag and kmers 21, 33, 55, 77 and 99. Contigs displaying less than 10-fold coverage were removed. Typing and identification of resistance markers for each isolate were assessed in silico (see Methods section in Supplementary Material).
SHRiMP (http://compbio.cs.toronto.edu/shrimp/; accessed 26 August 2019) as implemented in Nesoni v0.132 (https://github.com/Victorian-Bioinformatics-Consortium/nesoni; accessed 26 August 2019) was used with default settings to determine single nucleotide variants (SNVs) between the pre- and post-treatment isolates for each case, using their respective complete genome as a reference. Core SNVs were defined using Nesoni’s nway function with the ‘–require-all’ flag. SNVs associated with erroneous mapping of reads to repetitive regions and those found in regions of low coverage (<10-fold coverage) were subsequently removed. The Artemis Comparison Tool (http://sanger-pathogens.github.io/Artemis/ACT/; accessed 26 August 2019)26 was used to manually inspect read pileups and to detect structural variants.
For each case, short reads were also aligned using the same approach to both close and distantly related, publicly available N. gonorrhoeae reference genomes determined from a pairwise distance matrix (see Methods section in Supplementary Material). Genome coverage plots using reference genomes of varying genetic relatedness can be seen in Figure S1, available as Supplementary Material to this paper. A phylogeny comprising isolates from both cases and publicly available genomes was generated as described previously,18 and further details are given in the Results section of the Supplementary Material.
The number of core genes was determined using Roary v3.12 (https://sanger-pathogens.github.io/Roary/; accessed 26 August 2019)27 for de novo assembled isolates from Case 2 and included either the close or distant publicly available reference genome (see the Results section in the Supplementary Material).
Data availability
For Case 1, genome data have been deposited under BioProject PRJNA451380, which comprises the pre-treatment isolate TFG-B1 and post-treatment isolate TFG-B2 (GenBank accession CP032429). For Case 2, genome data have been deposited under BioProject PRJNA451379, which comprises pre-treatment isolate TFG-A1 and post-treatment isolate TFG-A2 (GenBank accession CP032398). A summary of methylation profiles for both TFG-B2 and TFG-A2 is given in Table S1, available as Supplementary Material to this paper.
Ethics approval
This study was approved by the South Eastern Sydney Local Health District Human Research Ethics Committee.
Results
Case 1
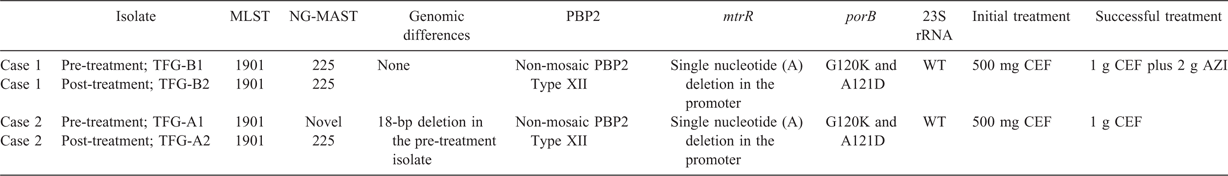
The overall TFG-B2 PacBio post-treatment genome size was 2 224 596 bp, which comprised a 2 220 291 bp chromosome and a 4305 bp cryptic plasmid. Both pre- and post-treatment isolates were MLST 1901 and NG-MAST 225. They both comprised a non-mosaic penicillin-binding protein 2 Type XII sequence, which is encoded by the penA gene, a single nucleotide (A) deletion in the promoter of the multiple transferable resistance Regulator, which is encoded by the mtrR gene, wild-type 23S rRNA alleles and two previously characterised non-synonymous nucleotide substitutions in porB (Table 1). No SNVs were detected between the pre- and post-treatment isolates after excluding base calls at both repetitive and low coverage regions in the TFG-B2 reference genome (Table S2). However, a comparison of the pre- and post-treatment draft genomes revealed the deletion of a 2.3 kb region involving the gonococcal pilin subunit encoded by pilE in the pre-treatment isolate. Loss of pilE in the pre-treatment isolate was confirmed using read mapping. Notably, one region excluded from the SNV analysis (positions 2110232–2110249 in TFG-B2) was located upstream of pilE, and is known to comprise portions of a partial pilin gene copy including a conserved region known as cys2 (Figure S2).28 The TFG-B1 SNVs identified at this region were well supported by read coverage (Table S2), suggesting this could be a genuine difference between isolates. However, highly conserved cys2 regions are also present immediately preceding the hypervariable tail encoded by the 3′ end of pilE and multiple silent copies encoded by pilS loci distributed in other parts of the genome, suggesting that mismapping accounts for these differences (Figure S3).
Case 2
The overall size of the TFG-A2 PacBio post-treatment genome was 2 225 966 bp, which comprised a 2 221 661 bp chromosome and a 4305 bp cryptic plasmid. Both isolates were MLST 1901, but had different NG-MAST profiles. The post-treatment isolate was identified as NG-MAST 225, but the pre-treatment isolate exhibited a novel NG-MAST profile due to the 18-bp deletion in porB. The isolates shared the same resistance markers as reported above for Case 1 (summarised in Table 1). Owing to the pilE observation for Case 1, pilE was also specifically interrogated. A 2.7 kb region that included pilE was found to be missing from the pre-treatment isolate, indicating a deletion of this gene. No other SNVs were detected between the pre- and post-treatment isolates after excluding calls at repetitive and low-coverage regions in the TFG-A2 reference genome (Table S3). Notably, one of the excluded regions (positions 76865–76969) corresponded to a phase variable repeat tract associated with the opacity-associated protein encoded by opa.
Additional testing
For Case 2, we also developed real-time polymerase chain reaction (PCR) assays to confirm the 18-bp deletion was present in the pre-treatment isolate (Table S4) because this deletion was previously reported in the post-treatment isolate.8 Briefly, we used both high-resolution melting and allele-specific PCR assays (Table S4) to detect the deletion. These PCR assays were applied to the DNA prepared for sequencing along with the original suspensions used in the initial investigation of these cases. This PCR testing confirmed a mistake had been made in the original article, and that the 18-bp deletion was in the pre-treatment isolate for Case 2. As a result, we are preparing a correction to Sexual Health.
Assessing the suitability of short-read sequence data for characterising cases of treatment failure
Because not all laboratories have the capacity to rapidly generate complete reference genomes on a case-by-case basis, we investigated whether short-read sequence data alone were sufficient for assessing treatment failures. Fifteen high-quality N. gonorrhoeae complete reference genomes were obtained from GenBank with two (32867 and FA1090; GenBank accessions CP016015.1 and AE004969.1 respectively) selected for further comparison based on their genetic distance to isolates from Case 1 and Case 2 (Figures S4, S5). Reference 32867 was found to be the most closely related strain to Case 1 and Case 2, differing by 312 and 315 single nucleotide polymorphisms (SNPs) respectively, and shares the same MLST and NG-MAST profile. Reference FA1090 (MLST 1899 and NG-MAST 773) was one of the most distantly related strains, differing from either case by over 4000 SNPs, and is from a different ancestral lineage.
Read mapping to reference 32867 identified minor variation between the pre- and post-treatment isolates from Case 1 and Case 2, differing by 1 and 21 SNVs respectively. When using the distantly related reference FA1090, the pre- and post-treatment isolates for Case 1 and Case 2 differed by 73 and 82 SNVs respectively. However, following the removal of SNVs associated with repetitive or low-coverage regions, pre- and post-treatment isolates from either case were found to be indistinguishable. It should be noted that variants called between pre- and post-treatment isolates from Case 2 include the 18-bp deletion in porB. The location of these SNVs for each case can be found in Tables S2 and S3 for Cases 1 and 2 respectively.
Phylogenetic context for the two treatment failure cases and other publicly available isolates
In the context of 15 complete N. gonorrhoeae reference genomes, the phylogeny revealed that the isolates in Case 1 were very closely related to isolates in Case 2, with only 28 nucleotide substitutions separating them (Figure S4). To further explore the relatedness between these cases, an additional phylogeny was constructed using a global collection of 66 publicly available N. gonorrhoeae clinical isolates (Table S5)17,19,29–31 with the same MLST (1901) and NG-MAST (225) profiles (Figure S6). The complete genome of N. gonorrhoeae 32867 was also included as a reference. The treatment failure cases cluster with 25 isolates (from several countries) in a distinct, highly supported subclade (86% bootstrap support) separated from the other isolates by a single defining nucleotide substitution. This was a non-synonymous substitution (K94N) in the EamA-like family transporter protein AS012_00330 (as annotated in reference genome 32867, base position 62729 of the complete sequence).
Discussion
In this study we compared pre- and post-treatment isolates from two previous cases (one confirmed and one suspected) of N. gonorrhoeae ceftriaxone treatment failure as examples to assess the feasibility and utility of WGS. Given the limited number of reports of N. gonorrhoeae ceftriaxone treatment failure for which pre- and post-treatment isolates have been available (only 13 cases documented to date),3,5–10,13 the isolates from these two cases provided a rather rare opportunity to assess the value of using WGS compared with NG-MAST. We also emphasise that having appropriate laboratory methods to help investigate potential cases of ceftriaxone treatment failure are of increasing importance, particularly as ceftriaxone resistance is continuing to emerge.4
For Case 1, for which both the previous clinical and NG-MAST data were consistent with treatment failure, no differences were detected between the two isolates using WGS, aside from a pilE deletion observed only in the pre-treatment isolate. This is consistent with previous findings elsewhere showing that multiple isolates taken from the same patient,17,18 or known sexual contacts,32 have very limited core differences. For Case 2, WGS confirmed the 18-bp deletion in porB (albeit in the pre- rather than post-treatment isolate; a correction is being prepared for the relevant journal regarding this mistake), but otherwise no other genomic differences were noted aside from a pilE deletion similar to that observed in Case 1. In the specific context of trying to confirm or exclude treatment failures, Case 2 highlights the benefits of using the enhanced resolution of WGS over NG-MAST by being able to compare the whole genome of two isolates, rather than two highly variable regions that undergo frequent antigenic variation. Of note is that there have only been five likely ceftriaxone treatment failure cases reported from Australia,5,7,8 all in the past 10 years, and in two of these cases (Case 2 herein and the case reported by Chen et al.7), changes in porB hindered NG-MAST interpretation. In both cases, additional DNA sequencing and phenotypic AMR results were compared to help interpret the NG-MAST data. We contend that such ad hoc investigations are far from ideal and that WGS offers a new opportunity to standardise laboratory protocols used for verifying gonorrhoea treatment failure.
Although WGS provides enhanced resolution, it is important to map read data to an appropriate reference genome to provide the most accurate comparison.33 Coverage plots can help assess regions where there is a significant drop in coverage, which are indicative of unreliable areas to call SNVs and often include repetitive regions. Selecting a distantly related genome will likely result in more false-positive SNVs and can reduce the likelihood of detecting true differences as the core genome becomes smaller (see Results section of the Supplementary Material). To generate an accurate SNV profile it is essential to filter out these false-positives,33 which are often the result of mismapping within repetitive regions. This can be achieved by masking these repetitive regions within the reference genome. Furthermore, there may be near-identical repetitive regions among the genomes of interest that are present in fewer copies in the reference genome, which will also cause false-positive SNVs. This was observed when using the distantly related strain (FA1090), indicating that the use of a closely related reference may reduce the likelihood of this issue arising. If further investigations of repeat regions are needed, either long-read sequencing or targeted resequencing can help explore these regions. However, even with the advantage of a closely related reference in this study, we still needed to exclude certain regions from analysis. For example, for Case 2 we excluded a region encompassing the phase variable repeat tract of the opa gene, for which there are several highly similar copies in the N. gonorrhoeae genome that are known to independently phase vary on or off depending on repeat tract length.34 Similarly, for Case 1, regions residing near pilE were excluded (positions 2110232–2110249). PilE contributes to pathogenesis via adhesion and is known to undergo high rates of antigenic variation due to recombination of multiple pilS loci into pilE, which comprises both hypervariable and highly conserved regions.35
Independent deletions of pilE in the pre-treatment isolates for both cases is also intriguing, because may it suggest a link between pilE and treatment failure. However, there is limited evidence of clinical isolates lacking pilE.36 Although high rates of antigenic variation are often associated with immune evasion, the loss of pilE could be a favourable adaptation because it may not be necessary to use pili once inside the host. Further studies are needed to explore these questions, which are outside the scope of the present study. Although PilE is involved in initial attachment to host cells, the capacity of pilE deletion mutants to cause urethral infection has been demonstrated previously.37 Swanson et al.38 have previously shown that both piliated and non-piliated colonies can be cultured from a single isolate. This could explain the pilE discrepancy for the cases of treatment failure, whereby non-piliated colonies were selected for when initially culturing these isolates. However, it is not possible to discern whether this loss occurred during infection or in vitro.
Overall, short-read sequencing was appropriate to help in the interpretation of these treatment failure cases. A critical advantage for sequencing treatment failures is that it allows us to rapidly assess relatedness to other cases. Although Cases 1 and 2 were identified within 5 months of each other in 2011 and shared a similar genotype and phenotypic profile, the original article treated them as two distinct events.8 However, the present study shows that the overall diversity between isolates in Cases 1 and 2 was minimal, clustering together in a distinct subclade with an additional 25 globally diverse MLST 1901 lineage isolates, within the broader MLST 1901 phylogeny. Surprisingly, this subclade was defined by a single non-synonymous substitution in the EamA-like family transporter protein AS012_00330. AS012_00330 is highly conserved within N. gonorrhoeae but its function is unknown. In Escherichia coli K12, a homologue of EamA is encoded by ydgE, which encodes for an efflux pump associated with the export of different metabolites of the cysteine pathway.39 AS012_00330 bears little resemblance to EamA from K12 (BLASTP 38% amino acid sequence identity, 14% sequence coverage). However, AS012_00330 does possess three transmembrane domains, suggesting it may be involved with the transport of specific substances across the membrane. Although it is tempting to speculate that the observed non-synonymous substitution in AS012_00330 may contribute to ceftriaxone treatment failures, there is no indication in the literature to suggest that the other 25 isolates that shared this mutation were, in fact, associated with treatment failure. Further work is warranted to determine the function of AS012_00330 and its contribution, if any, to N. gonorrhoeae ceftriaxone treatment failure.
NG-MAST 225 is prevalent here in Australia40 and elsewhere,41,42 with no further treatment failure cases having been reported with this sequence type. However, it is important to remain aware of potential cases, particularly those now failing dual therapy associated with azithromycin and ceftriaxone.3,13 However, this has become increasingly challenging with nucleic acid amplification tests now commonplace for diagnosing gonorrhoea.43 Therefore, only a minority of gonorrhoea infections have an appropriate culture (and associated phenotypic testing), suggesting there could be an underestimation of treatment failures, particularly in areas where culture data are either limited or non-existent.
Having now used WGS to confirm that the Case 2 isolates were indistinguishable (except for the porB and pilE deletions) raises further questions regarding Case 2 and the significance of porB. We are intrigued by the fact that the pre-treatment isolate harboured the 18-bp porB deletion and that this isolate was of a novel and presumably rare NG-MAST type. It is therefore somewhat counterintuitive that the pre-treatment isolate could then mutate into NG-MAST 225, which is a widely reported NG-MAST profile.40,44 Although the patient could have been reinfected before follow-up, they denied sexual intercourse during that period of time. While we still rely on individuals being truthful about when they engage in sexual intercourse, WGS provides enhanced resolution to aid in the investigation of treatment failures by being able to assess numerous genomic characteristics, including SNVs, structural variants and recombinant regions. This resolution is not achieved with traditional typing tools. We hypothesise that this patient may, in fact, have harboured a mixture of the above isolates (with and without the 18-bp porB deletion, and possibly the pilE deletion) before treatment, and that following treatment NG-MAST 225 was selected for. Unfortunately, we do not have the original swab samples available to test this hypothesis. Furthermore, although we only selected a single colony to culture, De Silva et al.17 have shown there is very minimal within-host diversity when independently sequencing a single anatomical site.
The 18-bp deletion is also intriguing in the context that the PorB protein is an essential outer membrane porin for maintaining N. gonorrhoeae viability that is subject to immune pressure and plays a key role in antimicrobial resistance;45 it is these factors that influence its high variability, particularly in the extracellular loops of the protein.46 Interestingly, the observed 18-bp deletion sits within extracellular loop 5, which likely (although not experimentally confirmed for this precise deletion) affects the binding of complement regulatory proteins, thereby conferring resistance to complement-mediated killing.47 Future experimental investigation is warranted to determine whether this precise deletion has any effect on ceftriaxone MIC. The changes in porB loop 3, involving amino acids G120 and A121, are well documented, and only appear to increase resistance in the presence of mtrR mutations.45,48 Although known resistance determinants affecting ceftriaxone were reported (porB and mtrR) for both cases, the only site to remain positive was the pharynx. This may suggest the primary reason for these ceftriaxone treatment failures is associated with suboptimal drug penetration into the oropharyngeal tissue.49
The WHO has indicated the need to develop standardised protocols for verifying N. gonorrhoeae treatment failure. Here, we propose that assessing the whole genome compared with two highly variable genes will provide both a more confident predictor for determining treatment failures and evidence of the immediate effect that WGS can have in an important and direct clinical application for N. gonorrhoeae. Although short-read sequencing is suitable for assessing these cases, it is important to use a closely related reference genome to reduce the number of false-positive SNVs that need to be excluded and provides a more accurate comparison. Moving forward, sequencing these treatment failures can provide a wealth of genomic information, including key resistance genes, and facilitate rapid comparison with similar cases elsewhere. Where available, we recommend using WGS for all suspected N. gonorrhoeae treatment failures.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors thank Tiffany Hogan and Rodney Enriquez from Prince of Wales Hospital (Sydney, NSW, Australia) and Helen Smith and Amy Jennison (Queensland Health Forensic and Scientific Services, Coopers Plains, Qld, Australia) for their assistance with the study. This work was supported by the Australian Infectious Diseases Research Centre and the National Health and Medical Research Council (GNT1090456 and GNT1105128 to Scott A. Beatson and David M. Whiley respectively).
References
[1] Unemo M, Del Rio C, Shafer WM. Antimicrobial resistance expressed by Neisseria gonorrhoeae: a major global public health problem in the 21st century. Microbiol Spectr 2016; 4| Antimicrobial resistance expressed by Neisseria gonorrhoeae: a major global public health problem in the 21st century.Crossref | GoogleScholarGoogle Scholar | 27337478PubMed |
[2] Department of Health. Gonorrhoea health alert. Canberra: Australian Government; 2018. Available online at: https://www1.health.gov.au/internet/main/publishing.nsf/Content/ohp-gonorrhoea.htm [verified 25 August 2019].
[3] Public Health England. UK case of Neisseria gonorrhoeae with high-level resistance to azithromycin and resistance to ceftriaxone acquired abroad. Health Protection Report Advanced Access Report, Volume 12, Number 11. 2018. Available online at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/694655/hpr1118_MDRGC.pdf [verified 23 August 2019].
[4] Lahra MM, Martin I, Demczuk W, Jennison AV, Lee KI, Nakayama SI, et al Cooperative recognition of internationally disseminated ceftriaxone-resistant Neisseria gonorrhoeae strain. Emerg Infect Dis 2018; 24 735–43.
| Cooperative recognition of internationally disseminated ceftriaxone-resistant Neisseria gonorrhoeae strain.Crossref | GoogleScholarGoogle Scholar |
[5] Tapsall J, Read P, Carmody C, Bourne C, Ray S, Limnios A, et al Two cases of failed ceftriaxone treatment in pharyngeal gonorrhoea verified by molecular microbiological methods. J Med Microbiol 2009; 58 683–7.
| Two cases of failed ceftriaxone treatment in pharyngeal gonorrhoea verified by molecular microbiological methods.Crossref | GoogleScholarGoogle Scholar | 19369534PubMed |
[6] Unemo M, Golparian D, Hestner A. Ceftriaxone treatment failure of pharyngeal gonorrhoea verified by international recommendations, Sweden, July 2010. Euro Surveill 2011; 16 19792
| 21329645PubMed |
[7] Chen MY, Stevens K, Tideman R, Zaia A, Tomita T, Fairley CK, et al Failure of 500 mg of ceftriaxone to eradicate pharyngeal gonorrhoea, Australia. J Antimicrob Chemother 2013; 68 1445–7.
| Failure of 500 mg of ceftriaxone to eradicate pharyngeal gonorrhoea, Australia.Crossref | GoogleScholarGoogle Scholar |
[8] Read PJ, Limnios EA, McNulty A, Whiley D, Lahra MM. One confirmed and one suspected case of pharyngeal gonorrhoea treatment failure following 500 mg ceftriaxone in Sydney, Australia. Sex Health 2013; 10 460–2.
| One confirmed and one suspected case of pharyngeal gonorrhoea treatment failure following 500 mg ceftriaxone in Sydney, Australia.Crossref | GoogleScholarGoogle Scholar | 24028864PubMed |
[9] Unemo M, Golparian D, Potocnik M, Jeverica S. Treatment failure of pharyngeal gonorrhoea with internationally recommended first-line ceftriaxone verified in Slovenia, September 2011. Euro Surveill 2012; 17 20200
| 22748003PubMed |
[10] Golparian D, Ohlsson A, Janson H, Lidbrink P, Richtner T, Ekelund O, et al Four treatment failures of pharyngeal gonorrhoea with ceftriaxone (500 mg) or cefotaxime (500 mg), Sweden, 2013 and 2014. Euro Surveill 2014; 19 20862
| Four treatment failures of pharyngeal gonorrhoea with ceftriaxone (500 mg) or cefotaxime (500 mg), Sweden, 2013 and 2014.Crossref | GoogleScholarGoogle Scholar | 25108533PubMed |
[11] World Health Organization (WHO). WHO guidelines for the treatment of Neisseria gonorrhoeae. Geneva: WHO; 2016.
[12] Cole MJ, Spiteri G, Jacobsson S, Woodford N, Tripodo F, Amato-Gauci AJ, et al Overall low extended-spectrum cephalosporin resistance but high azithromycin resistance in Neisseria gonorrhoeae in 24 European countries, 2015. BMC Infect Dis 2017; 17 617
| Overall low extended-spectrum cephalosporin resistance but high azithromycin resistance in Neisseria gonorrhoeae in 24 European countries, 2015.Crossref | GoogleScholarGoogle Scholar | 28893203PubMed |
[13] Fifer H, Natarajan U, Jones L, Alexander S, Hughes G, Golparian D, et al Failure of dual antimicrobial therapy in treatment of gonorrhea. N Engl J Med 2016; 374 2504–6.
| Failure of dual antimicrobial therapy in treatment of gonorrhea.Crossref | GoogleScholarGoogle Scholar | 27332921PubMed |
[14] World Health Organization (WHO). Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. Geneva: WHO; 2012.
[15] Martin IM, Ison CA, Aanensen DM, Fenton KA, Spratt BG. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J Infect Dis 2004; 189 1497–505.
| Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area.Crossref | GoogleScholarGoogle Scholar | 15073688PubMed |
[16] Unemo M. Current and future antimicrobial treatment of gonorrhoea – the rapidly evolving Neisseria gonorrhoeae continues to challenge. BMC Infect Dis 2015; 15 364
| Current and future antimicrobial treatment of gonorrhoea – the rapidly evolving Neisseria gonorrhoeae continues to challenge.Crossref | GoogleScholarGoogle Scholar | 26293005PubMed |
[17] De Silva D, Peters J, Cole K, Cole MJ, Cresswell F, Dean G, et al Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect Dis 2016; 16 1295–303.
| Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study.Crossref | GoogleScholarGoogle Scholar | 27427203PubMed |
[18] Buckley C, Forde BM, Trembizki E, Lahra MM, Beatson SA, Whiley DM. Use of whole genome sequencing to investigate an increase in Neisseria gonorrhoeae infection among women in urban areas of Australia. Sci Rep 2018; 8 1503
| Use of whole genome sequencing to investigate an increase in Neisseria gonorrhoeae infection among women in urban areas of Australia.Crossref | GoogleScholarGoogle Scholar | 29367612PubMed |
[19] Harris SR, Cole MJ, Spiteri G, Sanchez-Buso L, Golparian D, Jacobsson S, et al Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic survey. Lancet Infect Dis 2018; 18 758–68.
| Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic survey.Crossref | GoogleScholarGoogle Scholar | 29776807PubMed |
[20] Didelot X, Dordel J, Whittles LK, Collins C, Bilek N, Bishop CJ, et al Genomic analysis and comparison of two gonorrhea outbreaks. MBio 2016; 7 e00525-16
| Genomic analysis and comparison of two gonorrhea outbreaks.Crossref | GoogleScholarGoogle Scholar | 27353752PubMed |
[21] Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 2013; 10 563–9.
| Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data.Crossref | GoogleScholarGoogle Scholar | 23644548PubMed |
[22] Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014; 9 e112963
| Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement.Crossref | GoogleScholarGoogle Scholar | 25409509PubMed |
[23] Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30 2068–9.
| Prokka: rapid prokaryotic genome annotation.Crossref | GoogleScholarGoogle Scholar | 24642063PubMed |
[24] Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30 2114–20.
| Trimmomatic: a flexible trimmer for Illumina sequence data.Crossref | GoogleScholarGoogle Scholar | 24695404PubMed |
[25] Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19 455–77.
| SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.Crossref | GoogleScholarGoogle Scholar | 22506599PubMed |
[26] Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis Comparison Tool. Bioinformatics 2005; 21 3422–3.
| ACT: the Artemis Comparison Tool.Crossref | GoogleScholarGoogle Scholar | 15976072PubMed |
[27] Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015; 31 3691–3.
| Roary: rapid large-scale prokaryote pan genome analysis.Crossref | GoogleScholarGoogle Scholar | 26198102PubMed |
[28] Hamrick TS, Dempsey JA, Cohen MS, Cannon JG. Antigenic variation of gonococcal pilin expression in vivo: analysis of the strain FA1090 pilin repertoire and identification of the pilS gene copies recombining with pilE during experimental human infection. Microbiology 2001; 147 839–49.
| Antigenic variation of gonococcal pilin expression in vivo: analysis of the strain FA1090 pilin repertoire and identification of the pilS gene copies recombining with pilE during experimental human infection.Crossref | GoogleScholarGoogle Scholar | 11283280PubMed |
[29] Demczuk W, Lynch T, Martin I, Van Domselaar G, Graham M, Bharat A, et al Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J Clin Microbiol 2015; 53 191–200.
| Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013.Crossref | GoogleScholarGoogle Scholar | 25378573PubMed |
[30] Demczuk W, Martin I, Peterson S, Bharat A, Van Domselaar G, Graham M, et al Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J Clin Microbiol 2016; 54 1304–13.
| Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014.Crossref | GoogleScholarGoogle Scholar | 26935729PubMed |
[31] Grad YH, Kirkcaldy RD, Trees D, Dordel J, Harris SR, Goldstein E, et al Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 2014; 14 220–6.
| Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study.Crossref | GoogleScholarGoogle Scholar | 24462211PubMed |
[32] Kwong JC, Chow EPF, Stevens K, Stinear TP, Seemann T, Fairley CK, et al Whole-genome sequencing reveals transmission of gonococcal antibiotic resistance among men who have sex with men: an observational study. Sex Transm Infect 2018; 94 151–7.
| Whole-genome sequencing reveals transmission of gonococcal antibiotic resistance among men who have sex with men: an observational study.Crossref | GoogleScholarGoogle Scholar | 29247013PubMed |
[33] Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, Bulach DM, et al Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 2016; 54 333–42.
| Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes.Crossref | GoogleScholarGoogle Scholar | 26607978PubMed |
[34] Meyer TF, Gibbs CP, Haas R. Variation and control of protein expression in Neisseria. Annu Rev Microbiol 1990; 44 451–77.
| Variation and control of protein expression in Neisseria.Crossref | GoogleScholarGoogle Scholar | 2252390PubMed |
[35] Seifert HS, Wright CJ, Jerse AE, Cohen MS, Cannon JG. Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J Clin Invest 1994; 93 2744–9.
| Multiple gonococcal pilin antigenic variants are produced during experimental human infections.Crossref | GoogleScholarGoogle Scholar | 7911129PubMed |
[36] Bilek N, Ison CA, Spratt BG. Relative contributions of recombination and mutation to the diversification of the opa gene repertoire of Neisseria gonorrhoeae. J Bacteriol 2009; 191 1878–90.
| Relative contributions of recombination and mutation to the diversification of the opa gene repertoire of Neisseria gonorrhoeae.Crossref | GoogleScholarGoogle Scholar | 19114493PubMed |
[37] Hobbs MM, Sparling PF, Cohen MS, Shafer WM, Deal CD, Jerse AE. Experimental gonococcal infection in male volunteers: cumulative experience with Neisseria gonorrhoeae strains FA1090 and MS11mkC. Front Microbiol 2011; 2 123
| Experimental gonococcal infection in male volunteers: cumulative experience with Neisseria gonorrhoeae strains FA1090 and MS11mkC.Crossref | GoogleScholarGoogle Scholar | 21734909PubMed |
[38] Swanson J, Bergstrom S, Barrera O, Robbins K, Corwin D. Pilus- gonococcal variants. Evidence for multiple forms of piliation control. J Exp Med 1985; 162 729–44.
| Pilus- gonococcal variants. Evidence for multiple forms of piliation control.Crossref | GoogleScholarGoogle Scholar | 2410533PubMed |
[39] Dassler T, Maier T, Winterhalter C, Bock A. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol Microbiol 2000; 36 1101–12.
| Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway.Crossref | GoogleScholarGoogle Scholar | 10844694PubMed |
[40] Whiley DM, Limnios EA, Ray S, Sloots TP, Tapsall JW. Diversity of penA alterations and subtypes in Neisseria gonorrhoeae strains from Sydney, Australia, that are less susceptible to ceftriaxone. Antimicrob Agents Chemother 2007; 51 3111–16.
| Diversity of penA alterations and subtypes in Neisseria gonorrhoeae strains from Sydney, Australia, that are less susceptible to ceftriaxone.Crossref | GoogleScholarGoogle Scholar | 17591846PubMed |
[41] Palmer HM, Young H, Graham C, Dave J. Prediction of antibiotic resistance using Neisseria gonorrhoeae multi-antigen sequence typing. Sex Transm Infect 2008; 84 280–4.
| Prediction of antibiotic resistance using Neisseria gonorrhoeae multi-antigen sequence typing.Crossref | GoogleScholarGoogle Scholar | 18256103PubMed |
[42] Monfort L, Caro V, Devaux Z, Delannoy AS, Brisse S, Sednaoui P. First Neisseria gonorrhoeae genotyping analysis in France: identification of a strain cluster with reduced susceptibility to Ceftriaxone. J Clin Microbiol 2009; 47 3540–5.
| First Neisseria gonorrhoeae genotyping analysis in France: identification of a strain cluster with reduced susceptibility to Ceftriaxone.Crossref | GoogleScholarGoogle Scholar | 19794054PubMed |
[43] Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 2014; 27 587–613.
| Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future.Crossref | GoogleScholarGoogle Scholar | 24982323PubMed |
[44] European Centre for Disease Prevention and Control. Molecular typing of Neisseria gonorrhoeae – results from a pilot study 2010–2011. Stockholm: ECDC; 2012.
[45] Olesky M, Hobbs M, Nicholas RA. Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob Agents Chemother 2002; 46 2811–20.
| Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae.Crossref | GoogleScholarGoogle Scholar | 12183233PubMed |
[46] Chen A, Seifert HS. Saturating mutagenesis of an essential gene: a majority of the Neisseria gonorrhoeae major outer membrane porin (PorB) is mutable. J Bacteriol 2014; 196 540–7.
| Saturating mutagenesis of an essential gene: a majority of the Neisseria gonorrhoeae major outer membrane porin (PorB) is mutable.Crossref | GoogleScholarGoogle Scholar | 24244002PubMed |
[47] Chen A, Seifert HS. Structure–function studies of the Neisseria gonorrhoeae major outer membrane porin. Infect Immun 2013; 81 4383–91.
| Structure–function studies of the Neisseria gonorrhoeae major outer membrane porin.Crossref | GoogleScholarGoogle Scholar | 24042111PubMed |
[48] Veal WL, Nicholas RA, Shafer WM. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J Bacteriol 2002; 184 5619–24.
| Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae.Crossref | GoogleScholarGoogle Scholar | 12270819PubMed |
[49] Lewis DA. Will targeting oropharyngeal gonorrhoea delay the further emergence of drug-resistant Neisseria gonorrhoeae strains? Sex Transm Infect 2015; 91 234–7.
| Will targeting oropharyngeal gonorrhoea delay the further emergence of drug-resistant Neisseria gonorrhoeae strains?Crossref | GoogleScholarGoogle Scholar | 25911525PubMed |